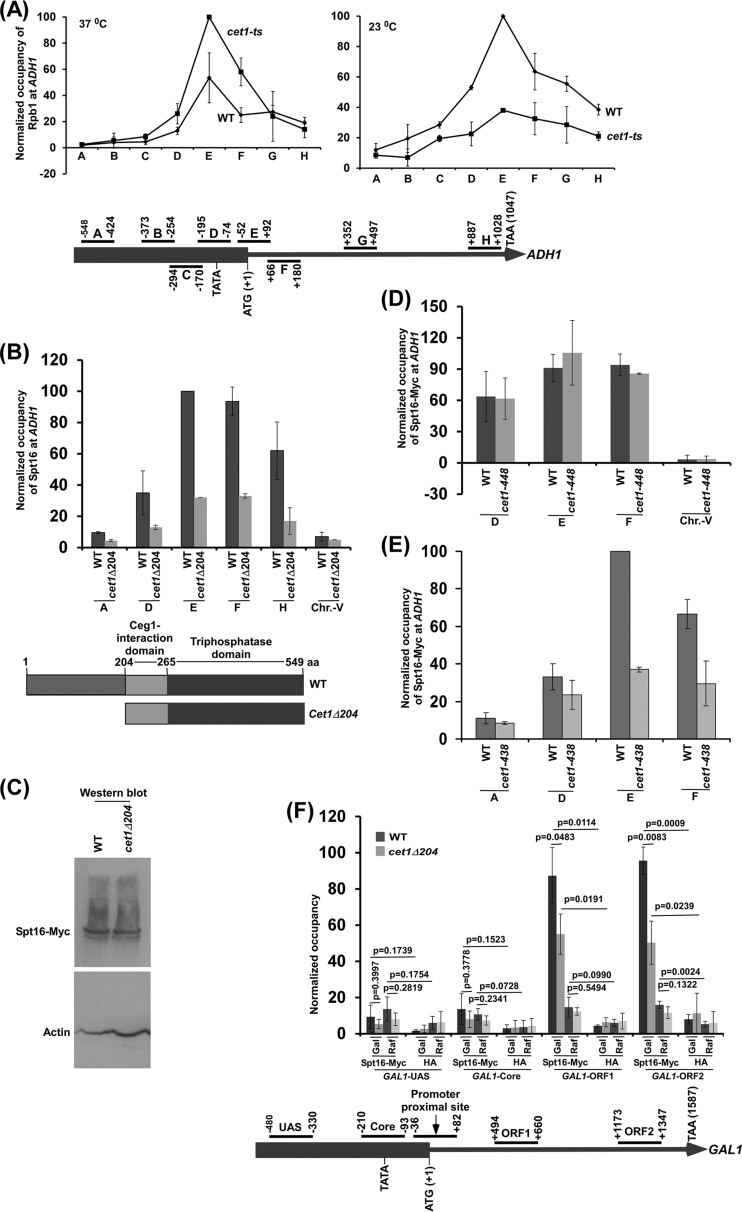
FIG 1.
The NTD (aa 1 to 204) of Cet1 targets FACT to ADH1 independently of mRNA-capping activity. (A) (Top) Analysis of Pol II association with ADH1 in the cet1(Ts) (cet1-438) mutant (YSB717) and wild-type (YSB540) strains at the nonpermissive (left) and permissive (right) temperatures. Immunoprecipitations were performed using mouse monoclonal antibody 8WG16 (Covance) against the CTD of Rpb1. The ratio of immunoprecipitate to input in the autoradiogram was measured. The maximum ratio was set to 100, and other ratios were normalized with respect to 100. The normalized ratio (represented as normalized occupancy) is plotted in the form of a histogram. (Bottom) Schematic diagram showing the locations of different primer pairs at ADH1 for ChIP analysis. The numbers are presented with respect to the position of the first nucleotide of the initiation codon (+1). (B) Analysis of FACT (Spt16-Myc) association with ADH1 in the presence and absence of the NTD of Cet1. (Top) Yeast cells were grown in YPD medium at 30°C to an OD600 of 1.0 prior to formaldehyde-based in vivo cross-linking. Immunoprecipitated DNA was analyzed by PCR using primer pairs targeted to different locations in ADH1. (Bottom) Schematic diagram of the different domains of Cet1. (C) Western blot analysis of Spt16 in the wild-type and cet1Δ204 strains. (D) ChIP analysis of FACT at ADH1 in the cet1(Ts) (cet1-448) mutant and its isogenic wild-type equivalent. Both wild-type and mutant cells were grown in YPD at 30°C to an OD600 of 0.85 and then switched to 37°C for 90 min prior to cross-linking. (E) ChIP analysis of FACT at ADH1 in the cet1(Ts) (cet1-438) mutant and its isogenic wild-type equivalent. Yeast cells were grown as for panel D. (F) ChIP analysis of Spt16 at GAL1 in the wild-type and cet1Δ204 strains in galactose (Gal)- or raffinose (Raf)-containing growth medium. The error bars indicate SD.