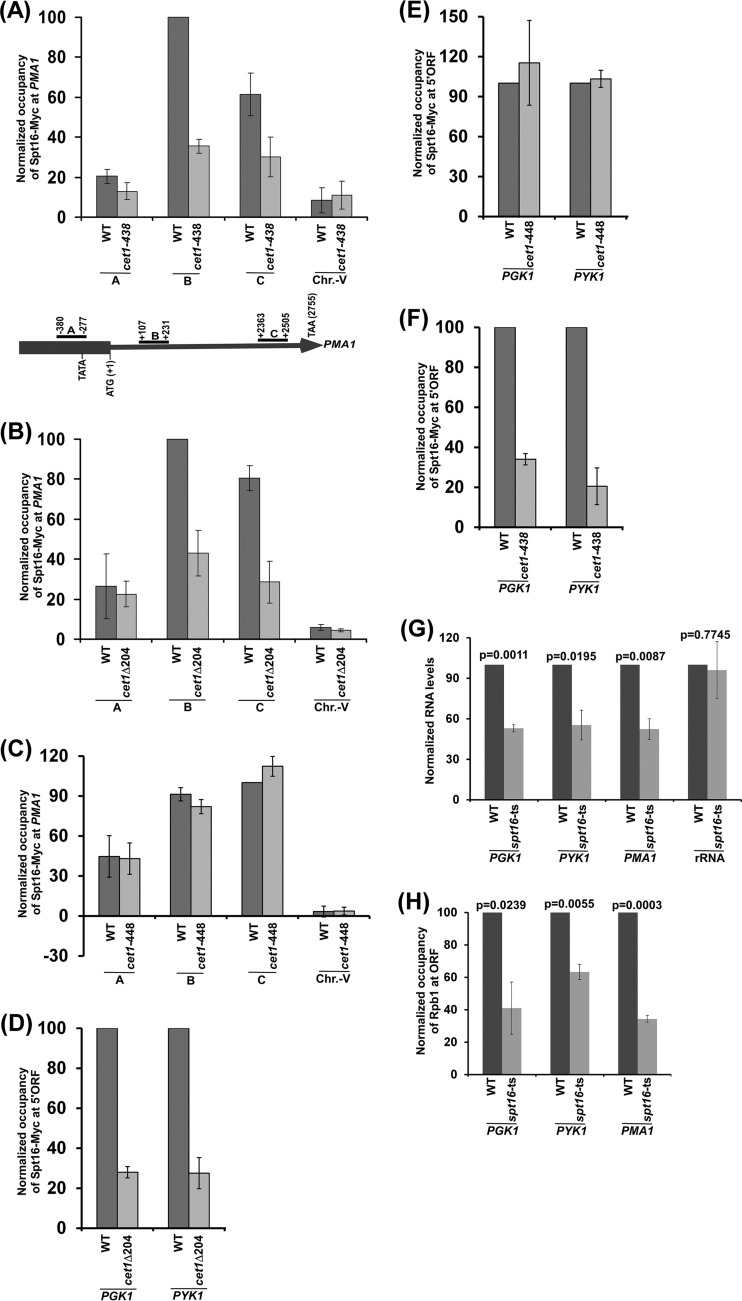
FIG 3.
The NTD of Cet1 targets FACT to PMA1, PGK1, and PYK1. (A) Analysis of FACT (Spt16-Myc) association with PMA1 in the cet1(Ts) (cet1-438) mutant and its wild-type equivalent. (B) Analysis of FACT (Spt16-Myc) association with PMA1 in the presence and absence of the NTD of Cet1. (C) Analysis of FACT (Spt16-Myc) association with PMA1 in the cet1(Ts) (cet1-448) mutant and its wild-type equivalent. (D) ChIP analysis of FACT at the 5′ ORFs of PGK1 and PYK1 in the presence and absence of the NTD of Cet1. (E) ChIP analysis of FACT at the 5′ ORFs of PGK1 and PYK1 in the cet1(Ts) (cet1-448) mutant and its wild-type equivalent. (F) ChIP analysis of FACT at the 5′ ORFs of PGK1 and PYK1 in the cet1(Ts) (cet1-438) mutant and its wild-type equivalent. (G) RT-PCR analysis of PGK1, PYK1, and PMA1 mRNAs in the spt16(Ts) mutant and its wild-type equivalent. (H) ChIP analysis of Rpb1 at the PGK1, PYK1, and PMA1 coding sequences in the spt16(Ts) mutant and its wild-type equivalent. The error bars indicate SD.