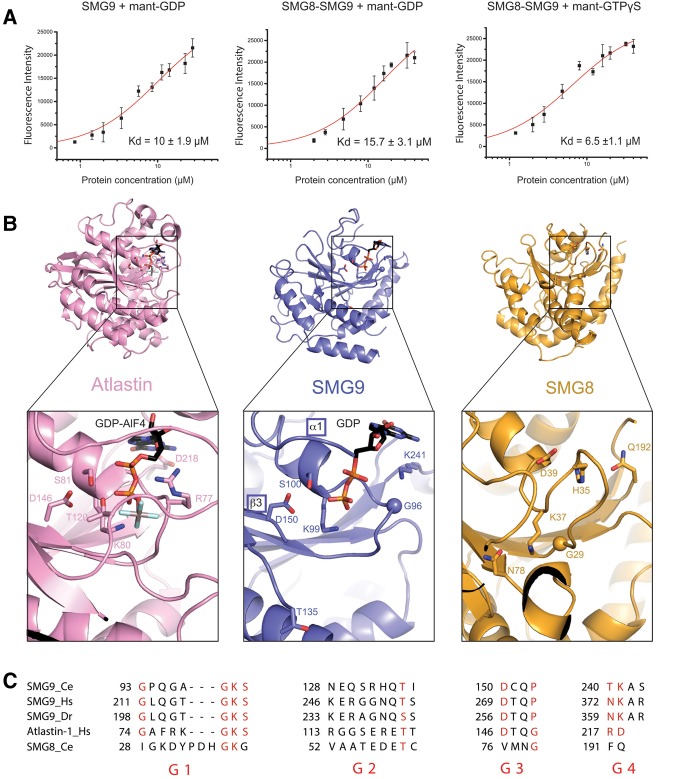
FIGURE 2.
The nucleotide-binding site of SMG9. (A) Fluorescence measurements of binding affinities of guanosine-nucleotides to SMG8c–SMG9c and SMG9c using mant-labeled GDP and GTP. The data were fitted to a binding equation describing a single-site binding model to obtain the dissociation constants (Kd). The best fit was plotted as a solid line. The Kd values and their corresponding errors are the mean and standard deviation of a minimum of three independent experiments. (B) Zoomed-in view at the nucleotide-binding site from the structure of SMG8c–SMG9c bound to GDP. The G domain of SMG9 is shown in the same orientation as in Figure 1B, left panel. The G domain of SMG8 and, as comparison, the G domain of Atlastin (bound to the GDP–AlF4 transition-state analog, ref) are shown in a similar orientation after optimal superposition. The nucleotides and important residues at the nucleotide-binding pockets of SMG9 and Atlastin are shown in ball-and-stick representation. Note that Thr135 in GDP-bound SMG9 (center panel) corresponds to Thr120 in GDP–AlF4-bound Atlastin (left panel). In SMG8, the equivalent site is incompatible with nucleotide binding: His35 and Gln192 would sterically clash with the ribose and base moieties, respectively, and Asp39 would lead to electrostatic repulsion with the phosphates. (C) Alignment of the G1–G4 motif sequences of SMG9 from C. elegans (Ce), H. sapiens (Hs), and D. rerio (Dr), and comparison with human Hs Atlastin and Ce SMG8. The position of the G motifs is schematized in Figure 1B: G1 (or P loop) in the β1–α1 loop, G2 (or switch 1) in α1–β2, G3 (or switch 2) in β3–α2, G4 in β5–α4, and G5 in β6–α5. The G5 motif is disordered in the present structure and divergent in sequence and therefore cannot be compared at present.