FIGURE 1.
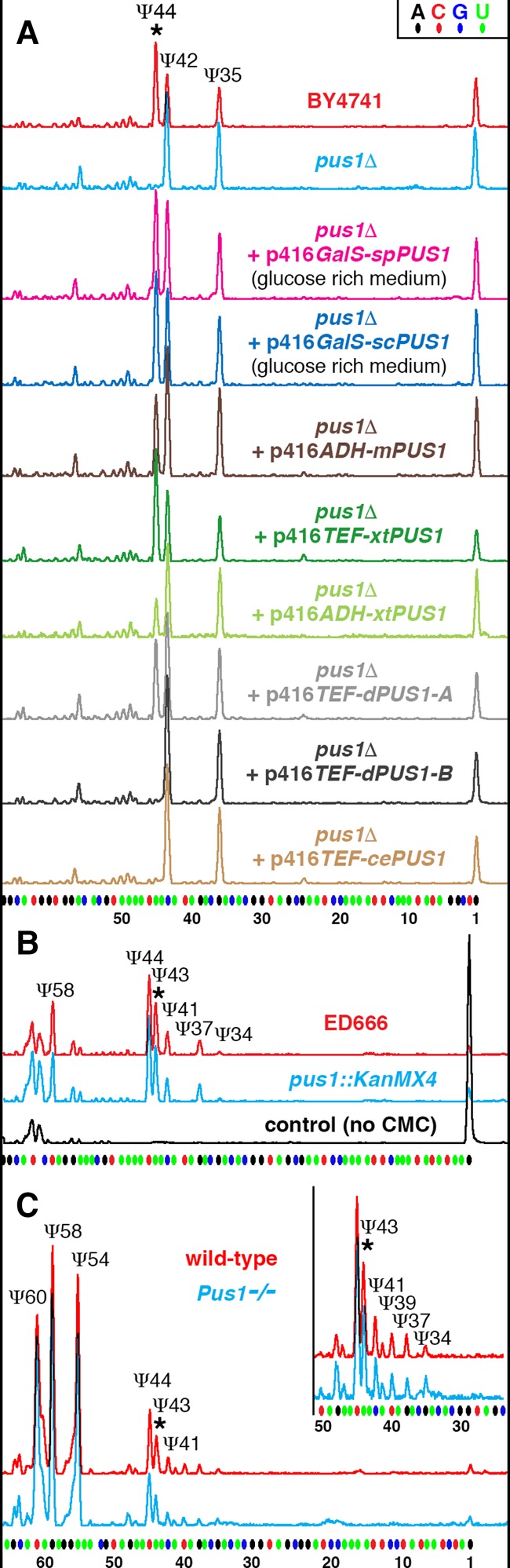
Pus1p enzymatic activity on U2 snRNA in different species. (A) S. cerevisiae U2 snRNA is normally pseudouridylated at positions 35, 42, and 44 (wild-type BY4741 strain, red trace). In a Pus1p-deficient S. cerevisiae strain, the pseudouridine at position 44 is missing (pus1Δ, blue trace). Pus1p from fission yeast S. pombe (spPus1p, magenta trace) when expressed in S. cerevisiae pus1Δ strain modifies yeast U2 snRNA as efficiently as S. cerevisiae Pus1p (scPus1p, dark blue trace). Mouse, Xenopus, and Drosophila Pus1p enzymes can rescue pseudouridylation at position 44 in yeast S. cerevisiae pus1Δ strain, yet a much higher level of expression is required (mPus1p, dark brown trace; xtPus1p, green traces; dPus1p-A/CG4159-PA, gray trace). Compare the efficiency of pseudouridylation when Xenopus Pus1p is expressed from plasmids with different promoter activities: ADH promoter (light green trace) and TEF promoter (dark green trace). Pus1p from C. elegans and the longer isoform of Drosophila Pus1p could not modify yeast U2 snRNA even when overexpressed (cePus1p, light brown trace; dPus1p-B/CG4159-PB, black trace). (B) U2 snRNA pseudouridylation mapping in S. pombe wild-type (ED666, red trace) and pus1Δ (pus1::KanMX4, blue trace) strains. U2 snRNA from the pus1Δ strain is modified at all the normal positions, including position 43. (C) U2 snRNA pseudouridylation mapping in wild-type (red trace) and Pus1 knockout (Pus1−/−, blue trace) mice. Inset in C zooms in on the branch point recognition region; note no differences between wild-type and mutant strains in their U2 snRNA modification patterns. Stars indicate peaks corresponding to pseudouridine at position 44 in S. cerevisiae U2 snRNA or equivalent position 43 in S. pombe and mouse U2 snRNA.
