FIGURE 1.
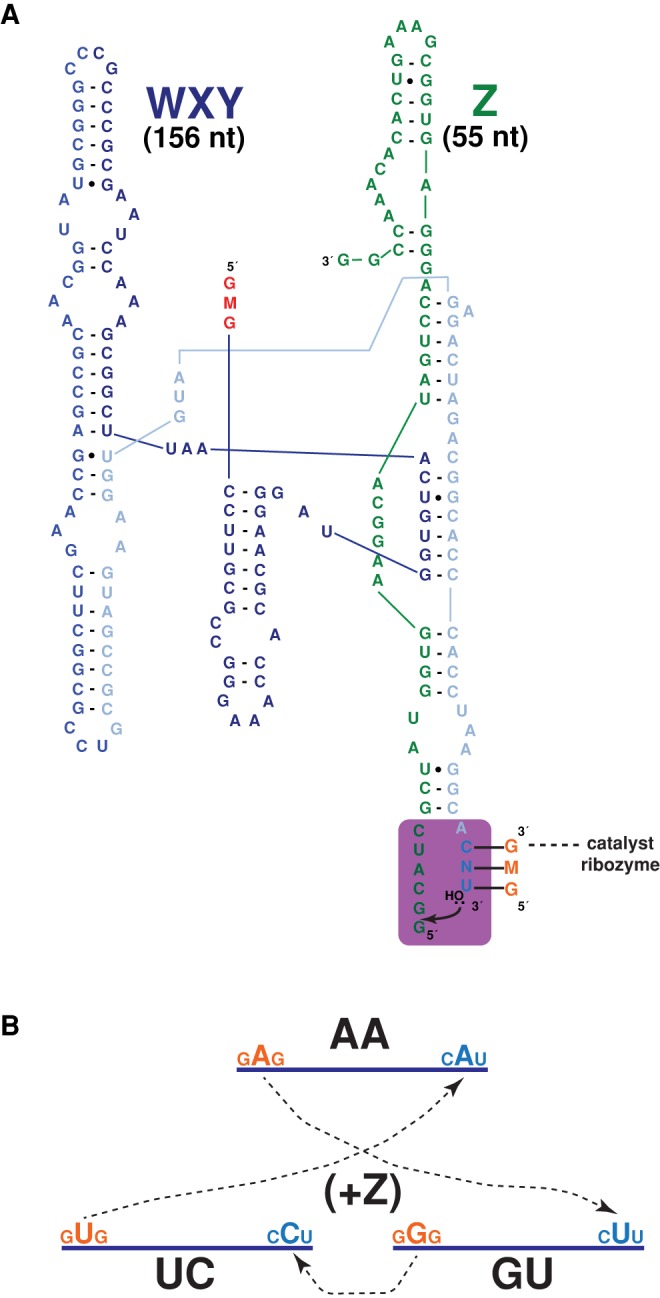
The Azoarcus ribozyme covalent self-assembly network. (A) The secondary structure of the ribozyme fragments and the catalytic event of the WXY+ Z → WXYZ reaction. The internal guide sequence (IGS) of the ribozyme lies at the 5′ end and is GMG, where M is free to vary among the four possible nucleotides. The IGS of one catalyst (symbolized by the “catalyst ribozyme”; it can be a covalently contiguous WXYZ or a noncovalent binding of a WXY to a Z) binds to the terminal 3 nt (CNU) of the WXY fragment in another molecule and catalyzes the recombination reaction shown, liberating a free G nucleotide and joining the substrate WXY with the Z. This reaction works at maximal rate if M and N form a Watson–Crick pair. (B) An example of the three-membered network possible if the M and N nucleotides are not complementary within a particular WXY molecule. Here the WXY species are symbolized by blue lines, and each can be denoted by just specifying these M and N nucleotides, such that this network is AA + UC + GU, which form the RPS core described below. The self-assembly of the network requires the presence of the Z substrate, indicated by the “+Z” in the center of the diagram.
