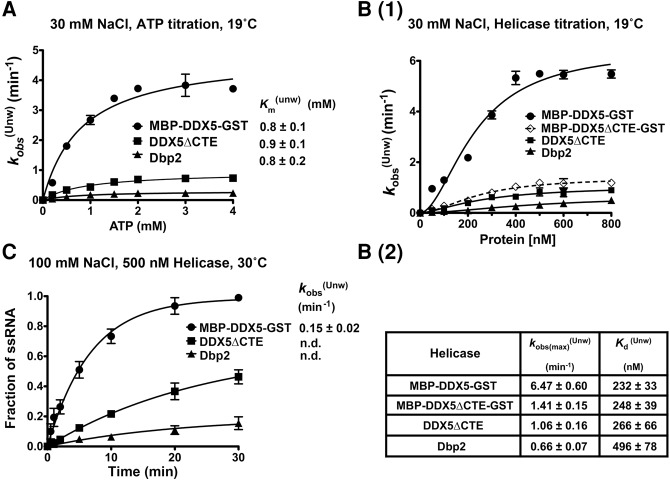
FIGURE 2.
The CTE confers higher duplex unwinding activity to human DDX5 over S. cerevisiae Dbp2. (A) MBP-DDX5-GST, DDX5ΔCTE, and Dbp2 have identical functional Km’s for ATP in unwinding assays. RNA unwinding assays were conducted using 300 nM recombinant, purified proteins and the indicated amount of ATP/MgCl2 in the presence of 30 mM NaCl at 19°C. The curves for observed unwinding rates versus ATP were fitted to a binding isotherm: Y = Kobs(max) × X/Km(unw) + X. (B) DDX5 shows higher unwinding activity than DDX5ΔCTE or Dbp2 in “low-salt” conditions. (1) Helicase unwinding assays were conducted at 19°C in the presence of 30 mM NaCl with indicated concentrations of purified recombinant MBP-DDX5-GST, DDX5ΔCTE, Dbp2, or MBP-DDX5ΔCTE-GST. (2) Table of maximum observed unwinding rates and the functional dissociation constants. First, the observed rates (kobs(Unw)) were determined by plotting the fraction of single-stranded (ss) RNA against reaction time and fitting the unwinding curves to the following equation: Y = Ymax × [1 − exp(−kobs(Unw) × X)]. To calculate the maximum observed rates (Kobs(max)(Unw)) and functional dissociation constants (Kd(Unw)), we plotted the observed rates as a function of the protein concentration and fitted the data to a sigmoidal binding isotherm [Y = Kobs(max)(Unw) × Xh/(Kd(Unw)h + Xh)] to generate functional binding curves. The Hill coefficients for these enzymes range from 1.5 to 1.8. (C) DDX5 is an active helicase in “near-physiological salt” conditions. Unwinding assays using 500 nM purified recombinant MBP-DDX5-GST, DDX5ΔCTE, or Dbp2 were performed in the presence of 100 mM NaCl at 30°C. The observed rates were determined as above. n.d., means not determined, when unwinding curves were not convergent. Data above show the mean ± standard deviation (SD) of 2–3 independent replicates.