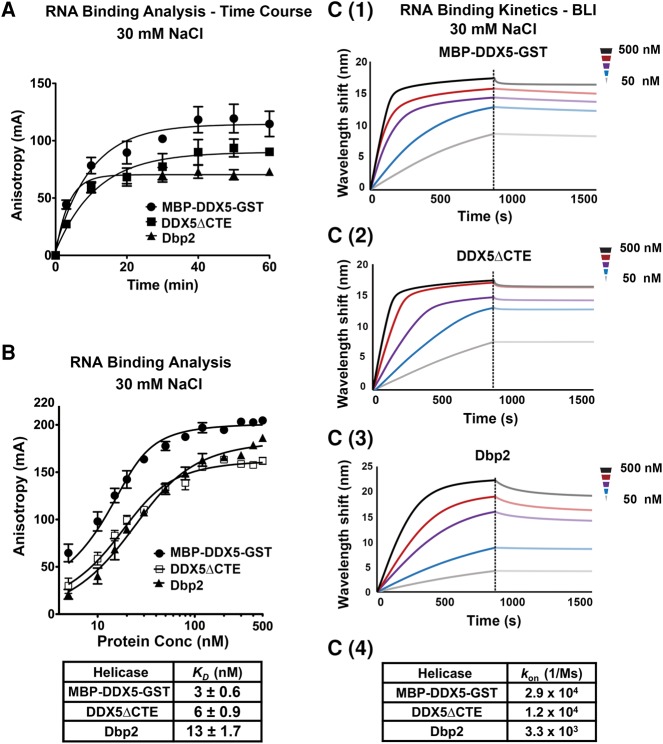
FIGURE 4.
Comparison of RNA-binding affinities and RNA on rates reveals tighter RNA binding by DDX5. (A) RNA-binding reactions reach equilibrium within 60 min. Fluorescence anisotropy assays were conducted using 20 nM indicated proteins, 10 nM 5′-6-FAM labeled 16-nt ssRNA, and AMPPNP. Anisotropy measurements (λex = 495 nm/λem = 520 nm) were taken using a Biotek Synergy 4 plate reader at indicated time points. (B) DDX5, DDX5ΔCTE, and Dbp2 bind ssRNA within a similar range, but with full-length DDX5 binding slightly tighter than DDX5ΔCTE and Dbp2. Fluorescence anisotropy assays were conducted using increasing amounts of MBP-DDX5-GST, DDX5ΔCTE, or Dbp2, 10 nM 16-nt ssRNA, and AMPPNP. Binding reactions were incubated for 60 min before taking anisotropy measurements. Binding data were fitted to the quadratic equation for two site binding: Y = Bmin + (Bmax − Bmin) × ((X + 2[RNA] + KD) − ((X + 2[RNA] + KD)2−8 × X × [RNA])0.5)/4. Data show the mean ± SD of three independent replicates. (C) DDX5 and DDX5ΔCTE show higher on rates than Dbp2 for ssRNA. Bio-layer interferometry (BLI) experiments were conducted to determine the binding kinetics of MBP-DDX5-GST (C1), DDX5ΔCTE (C2), and Dbp2 (C3) on a biotinylated 16-nt ssRNA in the presence of the nonhydrolyzable ATP analog, AMPPNP. Data are shown in background-corrected sensograms depicting the association and dissociation of proteins onto the ssRNA across a range of concentrations. The on-rates of MBP-DDX5-GST, DDX5ΔCTE, and Dbp2 were determined by fitting the real-time binding data with the 2:1 model using the Data Analysis Software (Pall Fortebio) by local fitting (C4). Off rates cannot be determined from this experiment due to the slow disassembly of DEAD-box helicase-AMPPNP–RNA complexes (Liu et al. 2014).