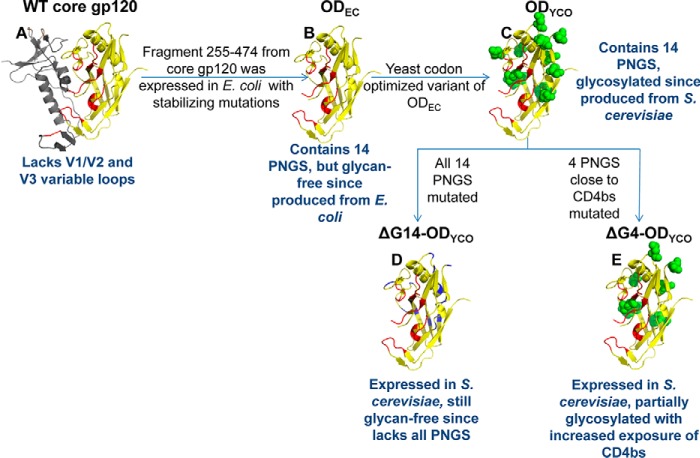
Figure 3.
Schematic representations of the glycosylation site mutants of OD fragments. A, structure of WT core gp120 (PDB code 1G9M). The OD fragment from residues 255 to 474 is shown in yellow; the CD4-binding site (CD4bs) is shown in red; and the rest of the protein is shown in gray. OD constructs (B–E) represent models of ODEC (all 14 PNGS are intact but still glycan-free due to expression in E. coli) (66), ODYCO (all 14 PNGS are intact, same amino acid sequence as ODEC but codon-optimized for expression in yeast, glycosylated due to expression in S. cerevisiae), ΔG14-ODYCO (devoid of all 14 PNGS and therefore glycan-free despite expression in S. cerevisiae), and ΔG4-ODYCO (four PNGS close to CD4bs mutated in ODYCO background, partially glycosylated), respectively. Asn residues at PNGS in C and E are shown as green spheres. Asn residues at PNGS mutated to other amino acids are shown in blue in D. These constructs are further described in Table 1.