Figure 9.
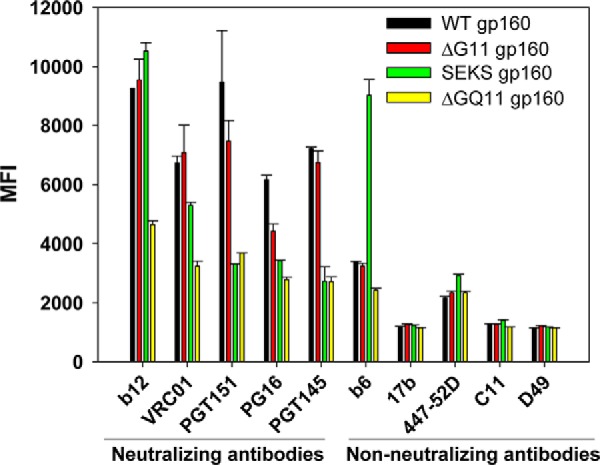
Effect of rationally designed versus Asn → Gln glycosylation site mutations on HIV-1 Env conformation probed by binding of anti-HIV-1 neutralizing and non-neutralizing mAbs to mammalian cell surface-expressed HIV-1 Env variants. WT gp160 and ΔG11 gp160 show similar binding profiles to CD4bs targeting neutralizing mAbs b12 and VRC01 as well as to the trimer-specific neutralizing antibodies PG16, PGT145, and PGT151. In contrast to the designed mutations, conventional Asn → Gln mutations at outer domain glycosylation sites (ΔGQ11 gp160) result in a significantly reduced binding of all neutralizing antibodies tested. Binding of ΔGQ11 gp160 with PGT145 and PGT151 is comparable with that of cleavage-defective SEKS gp160. A representative FACS histogram overlay for the binding of PGT151 antibody is shown in supplemental Fig. S14. A marginal decrease in the PGT151 and PG16 binding of rationally designed ΔG11 gp160 relative to WT gp160 could be attributed to the fact that binding of both these trimer-specific antibodies is dependent on the presence of certain core gp120 glycans (56, 86, 90). Both WT gp160 and ΔG11 gp160 show minimal binding to a number of different non-neutralizing mAbs with epitopes in diverse regions of the Env structure. In contrast, the cleavage-defective SEKS gp160 molecule shows high binding with the non-neutralizing mAb b6 but binds poorly to the trimer-specific neutralizing mAbs PGT145 and PGT151. All mAbs were used at a concentration of 1 μg/ml. Error bars represent standard deviation for the data from two independent experiments.
