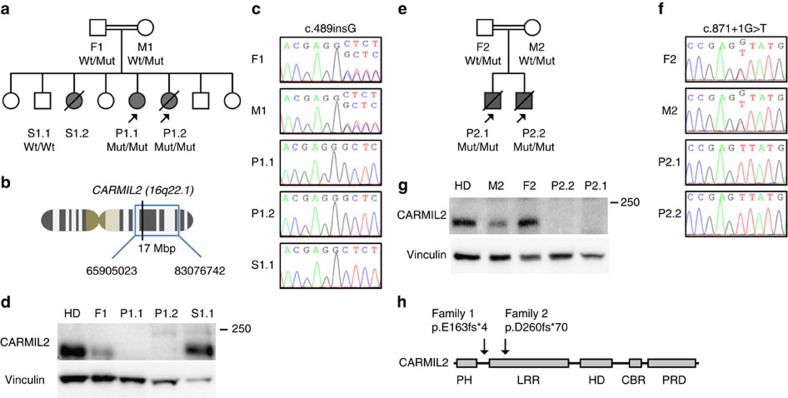
Figure 2. Homozygous CARMIL2 mutations segregate with the disease phenotype.
(a) Pedigree of family 1 with father (F1), mother (M1), siblings (S1.1 and S1.2) and patients (P1.1 and P1.2). Grey symbols and diagonal bars indicate diseased and deceased subjects, respectively, and CARMIL2 wildtype (Wt) and CARMIL2 c.489insG mutated (Mut) alleles are depicted for each patient. (b) Schematic representation of chromosome 16, cytogenetic band 16q22.1 and the homozygous region (blue box and bp interval) identified by SNP chip that harbour CARMIL2 (black vertical line). (c) Electropherograms of family 1 members for CARMIL2 Wt/Mut, Mut/Mut and Wt/Wt alleles. (d) CARMIL2 immunoblots of healthy donor (HD) and family 1 members with vinculin loading control. (e) Pedigree of family 2 with father (F2), mother (M2) and patients (P2.1 and P2.2). (f) Electropherograms of family 2 members for CARMIL2 Wt/Mut and Mut/Mut. (g) CARMIL2 immunoblot of HD and family 2 members with vinculin loading control. (h) Schematic representation of CARMIL2 protein domain architecture and localization of family 1 p.E163fs*4 and family 2 p.D260fs*70 mutations. Immunoblots in (d) and (g) have been repeated three times.