Figure 2.
NKX6-2 Mutations
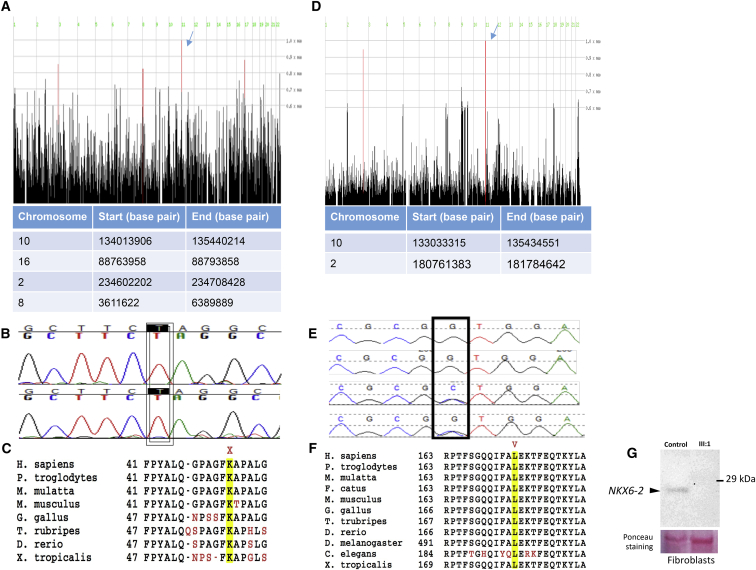
(A) Homozygosity mapping in families 1 and 2 identified four homozygous regions that were shared by the three affected individuals from families 1 and 2. The region on chromosome 10 includes NKX6-2 (arrow) shared between the two families.
(B) Sanger sequencing confirming c.121A>T in the two families.
(C) Conservation of p. Lys41∗ across species.
(D) Homozygosity mapping in family 3 identified two homozygous regions that were shared by the affected individuals. The region on chromosome 10 includes NKX6-2 (arrow).
(E) Sanger sequencing confirming c.487C>G and segregation in the family.
(F) Conservation of the p.Leu163Val residue within the NKX6-2 homeodomain across species. The mutation is marked in red above the corresponding amino acid.
(G) Immunoblot analysis showing a complete absence of NKX6-2 in an affected individual (individual III-1 in family 1 in the pedigree) compared to a control individual.