Fig. 2.
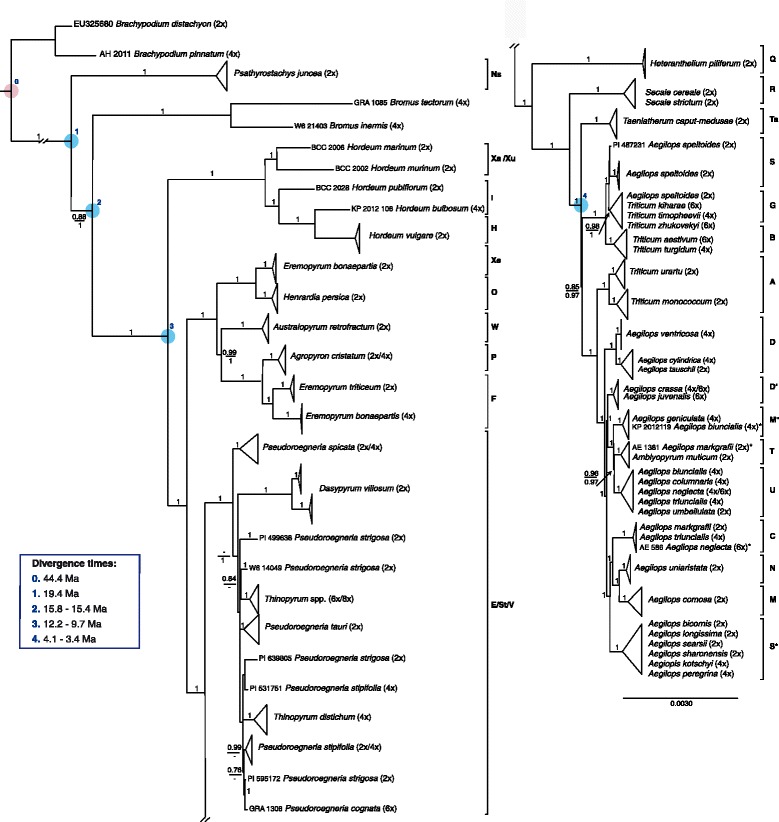
Phylogenetic tree derived from an alignment of whole genome chloroplast sequences via Bayesian phylogenetic inference. The multiple sequence alignment comprised 183 genomes assembled in the present study and 39 genomes that were downloaded from GenBank. Brachypodium distachyon was defined as outgroup taxon. The tree shown corresponds to an analysis based on the complete alignment of 123,531 base pairs (bp). Clades were collapsed into triangles to reflect the main groupings. The area of the triangles reflects the genetic variation contained in a certain clade. Posterior probabilities (pp) for the main clades are depicted next to the nodes if they were higher then 0.75. Support values of a second Bayesian phylogenetic analysis based on 114,788 bp of whole chloroplast genomes, where alignment positions with more than 50% of missing data were masked, are shown below the values of the corresponding nodes in the complete chloroplast analysis if the values differed between analyses. Ploidy levels are provided in brackets after the taxon labels. Single accessions grouping apart from other accessions of their taxon are highlighted with an asterisk. To the right the genomic groups are indicated. The red circle represents the secondary calibration point from Marcussen et al. [20] used for node calibrations in multispecies coalescent analyses (MSC). Major nodes are shown in blue and their estimated ages in million years are given in the box. Two age values for the same node correspond to the analysis with (first value) and without the inclusion of Psathyrostachys (second value). For more information on the results of the MSC analyses see Additional file 5: Figure S2 and Additional file 6: Figure S3. For the full representation of the tree showing the grouping of all single accessions see Additional file 4: Figure S1. For species synonyms see Additional file 1: Table S1. Arrows with support values indicate the nodes they refer to
