Figure 4. Recognition of short tandem repeat arrays by the DIM-5/DIM-2 pathway.
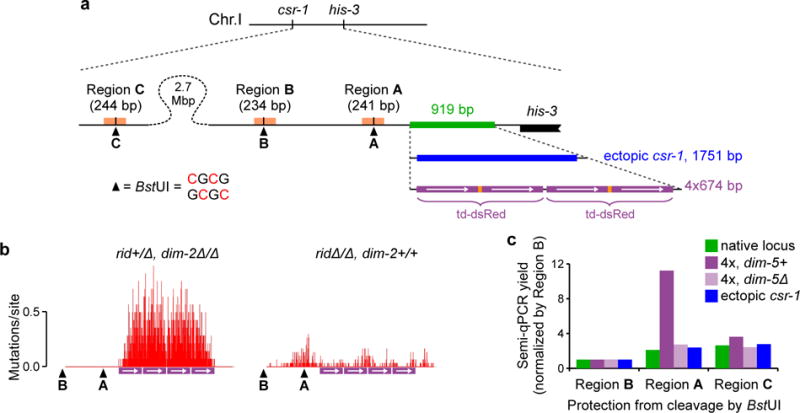
a, Four copies of the dsRed coding sequence (the “4x” array) are inserted by homologous recombination as the replacement of an endogenous genomic segment (green). The ectopic csr-1 copy is inserted analogously. All DNA segments are drawn to the same scale. The restriction site 5′-CGCG-3′ can only be cut by BstUI if none of its four cytosines is methylated (ref. 29). The distal BstUI site “C” is located near the csr-1 gene.
b, RIP mutation profiles of the 4x array. Crosses X27{14} (left) and X28{30} (right). The mean number of mutations in the entire sequenced region is 174.93± 30.84 and 15.20±1.69 for X26 and X27, respectively. Positions of the two proximal BstUI restriction sites (“A” and “B”, as shown in a) are indicated.
c, The assay detects the failure of cleavage by BstUI at the three specified sites (“A”, “B” and “C”, as shown in a). PCR yields for Regions A and C are normalized by the corresponding PCR yield for Region B. Unnormalized PCR yields are also provided in Supplementary Fig. 6b. The analyzed repeat constructs were never subjected to RIP. Strains: FGSC#9720 (native locus), T485.4h (4x, dim-5+), T486.3h (4x, dim-5Δ) and T402.1h (ectopic csr-1).
