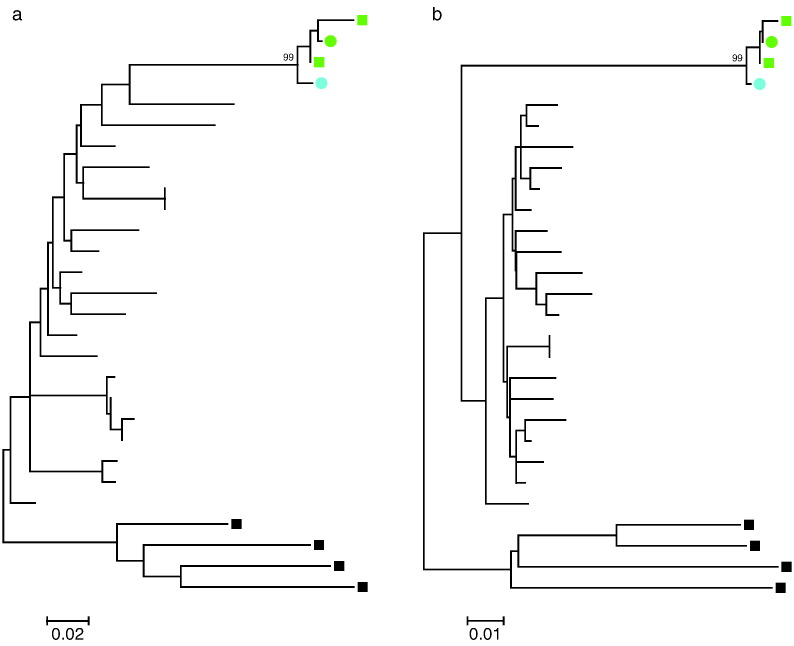
Fig. 3.
Phylogenetic analysis of viral sequences obtained from AC1 and VP1. Maximum likelihood phylogenetic trees of nef (a) and gag (b) nucleotide sequences are shown. Sequences amplified from replication-competent virus (circles) and provirus (squares) for AC1 (green) and VP1 (blue) are compared to the most homologous clade B sequences (branches without symbols) in the Los Alamos Database. Sequences from clade D (black squares) serve as an outgroup. Bootstrap values of the clades including AC1 and VP1 sequences are indicated.