Figure 3.
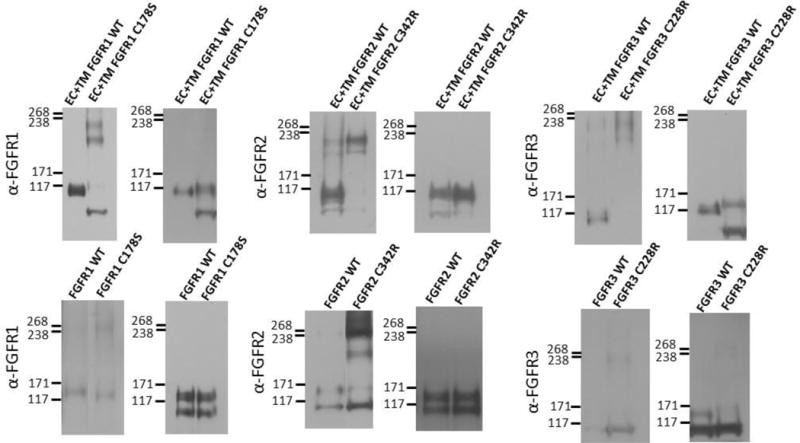
Anti-FGFR staining of (left) non-reducing and (right) reducing western blots for wild-type and mutant FGF receptors. Top row: truncated (EC+TM+YFP) receptors. Bottom row: full-length receptors. Typically, two monomeric and two dimeric bands can be observed, depending on the relative expression of the two forms, and the exposure. The lower bands correspond to immature, partially glycosylated receptors, while the higher molecular weight bands correspond to the mature, fully glycosylated forms of the receptors (~110 kDa for EC+TM+YFP and ~130 kDa for full length FGFR1–3)51–54. In these experiments, cells were starved in serum-free medium for 24 h following transfection with 1–3 μg of DNA encoding full length FGFRs. All experiments were performed with CHO cells, with the exception of the full-length FGFR3 blot, which was performed with HEK 293T cells due to poor expression of the C228 mutant in CHO cells. They were then treated with lysis buffer (25 mM Tris-HCl, 0.5% Triton X-100, 20mM NaCl, 2 mM EDTA, phosphatase inhibitor and protease inhibitor, Roche Applied Science). Lysates were collected following centrifugation at 15,000 g for 15 min at 4°C and loaded onto 3–8%NuPAGE®Novex®Tris-Acetatemini gels (Invitrogen, CA). The proteins were transferred onto a nitrocellulose membrane, and blocked using 5% milk in TBS. FGFR total protein levels were assessed using antibodies raised against the N-terminal epitope of FGFR3 (H-100; sc-9007), FGFR2 (H-80; sc-20735) and FGFR1 (H-76; sc-7945), all from Santa Cruz Biotechnology. This was followed by anti-rabbit HRP conjugated antibodies (W4011, Promega). The proteins were detected using the Amersham ECL detection system (GE Healthcare).
