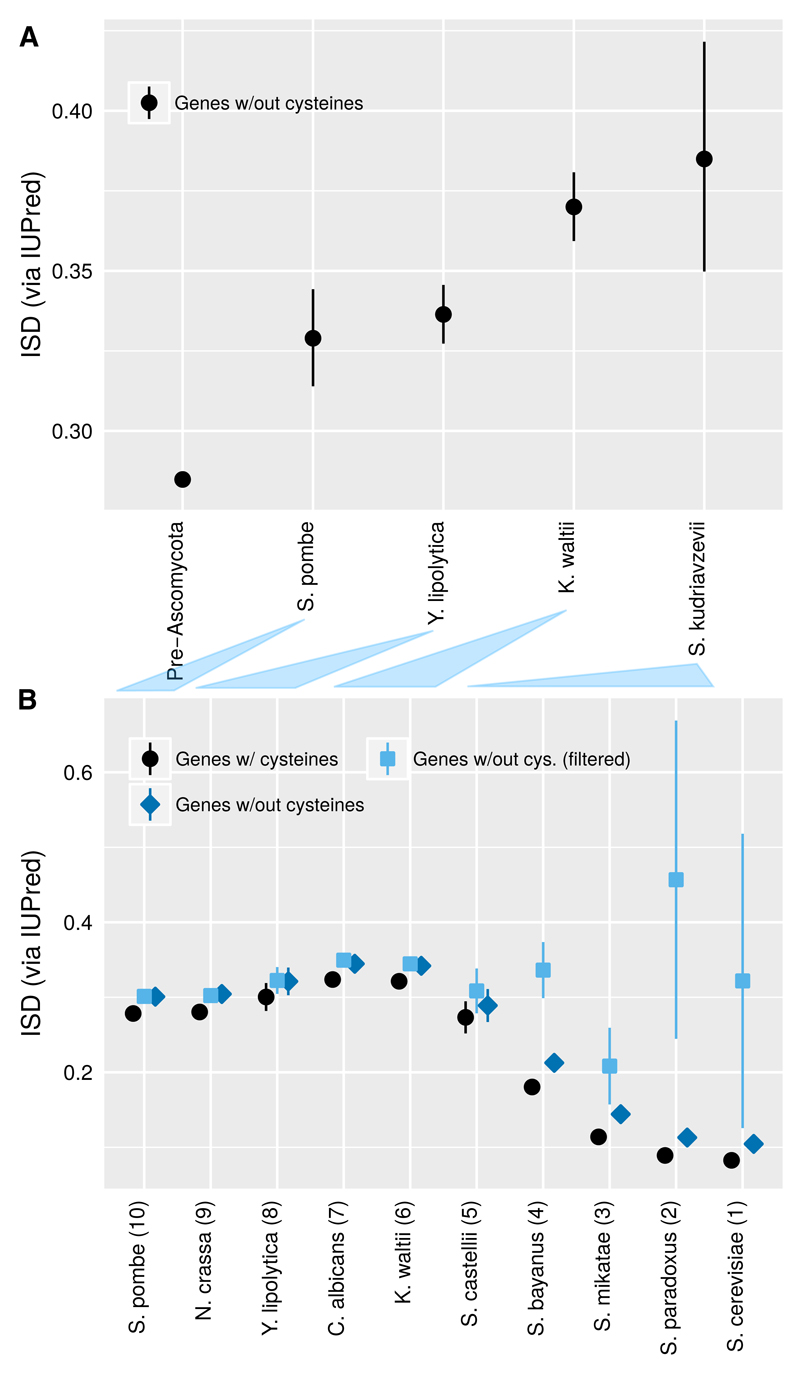
Fig. 6. Young yeast genes, like the young mouse genes in Fig. 2, have higher ISD.
A) Back-transformed central tendency estimates +/- one standard error come from a linear mixed model, where gene family and phylostratum are random and fixed terms, respectively. Phylostrata are labeled according to the species most closely related to S. cerevisiae in which a homolog is still found, except for the “S. kudriavzevii” group, which includes younger genes found in at least two species. The analysis includes 5452 yeast genes that overlap with the genes used by Carvunis et al. (2012) with filtering indicated in Table 1. B) Using the age classifications of Carvunis et al. (2012) (Table 1, 2nd column), and ignoring gene family, we reproduce the trend of low ISD in young “proto-genes” using our slightly different ISD measurement. Standard means +/- one standard error are reported for untransformed ISD estimates. This trend is insensitive to whether cysteines are included (black circles) or excluded (blue diamonds) from the protein primary sequence. This trend disappears when we screen out “proto-genes” that lack strong evidence for a functional protein product (light-blue squares), by excluding genes whose age we could classify or which were unique to S. cerevisiae, and those classified as “dubious” in SGD (Table 1; last column). Correspondences between the ages assigned by the two phylostratigraphies are indicated with shaded triangles between the two figure parts.