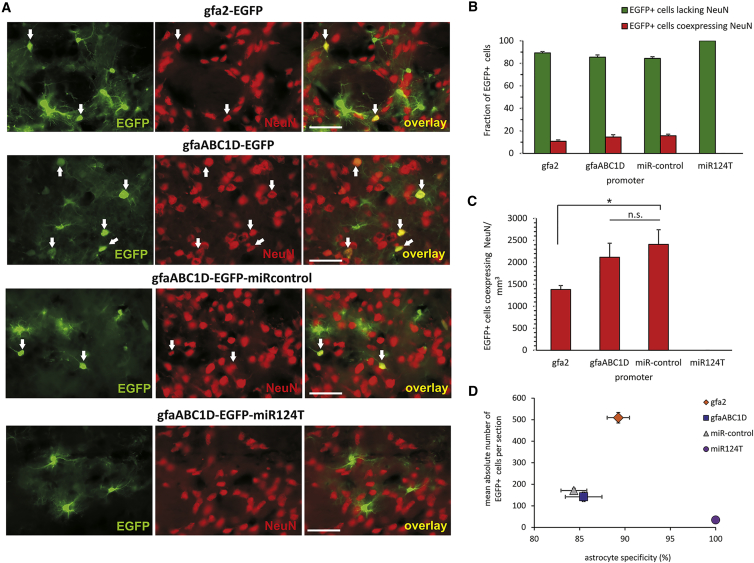
Figure 6.
Endogenous miR124 Effectively De-targeted Transgene Expression from Rat Neurons In Vivo
Immunohistochemical staining against NeuN was performed to test the interaction of neuronal miR124 and its targeting sequence in the EGFP 3′ UTR using constructs containing four copies of the artificial targeting sequence (miR124T) or reversed strand targeting sequence (miRcontrol) cloned into the 3′ UTR of the reporter gene. (A) Microscopy analysis of striatum of adult rats injected 3 weeks before with 1 × 108 tu of the indicated AAV6-EGFP vector. Counterstaining for NeuN (red) revealed EGFP fluorescence (green) in neurons. The merged images revealed some coexpressing cells. Shown are representative pictures from n = 3 rats per group. Arrows indicate EGFP+ neurons. Absence of any neuronal transgene expression by EGFP/NeuN overlay is shown for the AAV6-gfaABC1D-EGFP-miR124T vector. EGFP expression mediated by miR124T was limited exclusively to astrocytes. (B) Stereological quantification of transgene-expressing astrocytes and neurons. After insertion of miR124T sequences, EGFP+ neurons were not detected, and transgene expression was limited to astrocytes. (C) gfa2 and gfaABC1D caused a high level of neuronal expression. Only the gfaABC1D-miR124T viral vector was selective for striatal astrocytes. (D) The reporter transgene EGFP under control of the gfaABC1D promoter exhibited dramatic off-target, non-astrocytic expression patterns combined with a weak efficiency in vivo. Transgene expression restricted to astrocytes was restored by the incorporation of miR124T sequences. The gfa2 promoter was excellent in terms of activity, and EGFP immunoreactivity was mostly absent from neurons. Scale bars, 50 μm. *p = 0.038; p = 0.82, non-significant (n.s.). n = 5. One-way analysis of variance and post hoc Tukey’s test. Error bars represent SEM.