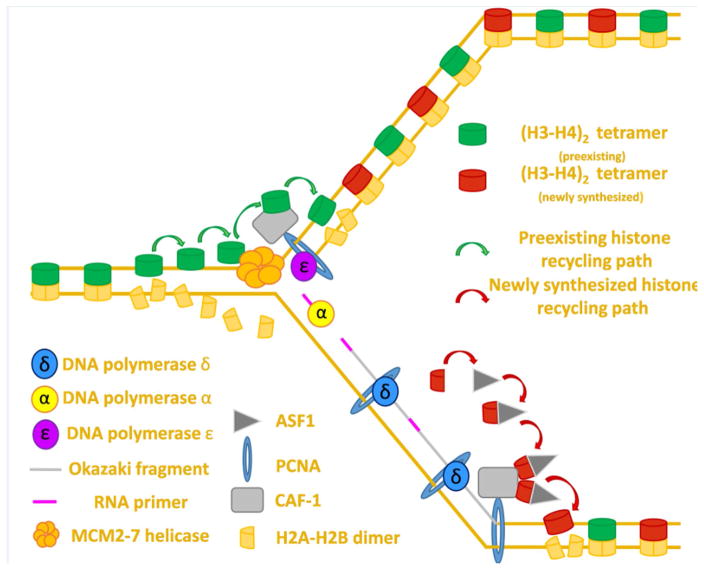
Figure 2. Proposed mechanism for directed histone recycling during DNA replication.
DNA replication overview: Polymerase ε synthesizes the leading strand continuously whereas polymerase δ synthesizes the lagging strand in discontinuous segments known as Okazaki fragments. Polymerase α synthesizes an RNA primer at the start of the leading strand (not shown) and at the start of each new Okazaki fragment.
Old histone recycling: The MCM2–7 helicase sits at the foremost edge of the replication fork where its primary function is to unwind non-replicated DNA. The MCM2 protein can bind (H3–H4)2 tetramers dissociated in the wake of the advancing fork to allow for their subsequent deposition on nascent DNA. Due to its ability to bind (H3–H4)2 tetramers, CAF-1 may help coordinate the deposition of preexisting histones after fork passage. CAF-1 is recruited to the edge of the advancing fork by PCNA, which also serves as a processivity factor for the replicative polymerases δ and ε.
New histone deposition: Histone chaperone ASF-1 binds (H3–H4) dimers in the cytoplasm and coordinates their import into the nucleus where ASF-1 transfers (H3–H4) dimers to CAF-1. CAF-1 then coordinates new H3–H4 deposition onto nascent DNA. The subsequent incorporation of two (H2A–H2B) dimers reforms the nucleosome structure and signifies the end of the process of replication coupled histone deposition.