Figure 1. Picornaviral Genome Structure and Polymerase Functions.
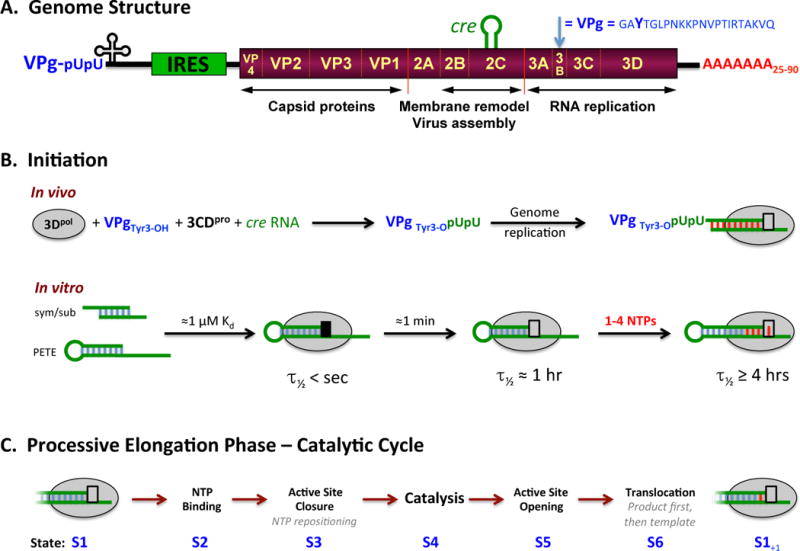
(A) Schematic representation of the poliovirus genome as a representative picornavirus. The genome encodes a single ≈250 KDa polyprotein that is translated from an internal ribosome entry site (IRES) and cleaved into about a dozen smaller proteins and functional intermediates by the viral 2Apro, 3Cpro, and 3CDpro proteases. The last part of the polyprotein is 3Dpol, a RNA-dependent RNA polymerase that is only active upon cleavage of the 3Cpro–3Dpol junction. Many picornaviruses replace 2Apro with a N-terminal leader (L) protease. (B) The native initiation pathway for 3Dpol uses the viral VPg protein (i.e. 3B) whose Tyr3 becomes doubly uridylylated via a cre RNA templated reaction in the context of a viral replication center. In vitro, however, 3Dpol will initiate using short RNA duplexes, such as the self-complementary sym/sub sequences used extensively by the Cameron lab or RNA hairpins “PETE” constructs used by the Peersen group. There is a stepwise assembly pathway whereby the initial 3Dpol-RNA complex needs to undergo a conformational transition to become catalytically competent, as indicated by the black versus grey box at the active site. (C) The full catalytic cycle that takes place repeatedly during processive elongation can be divided into six major structural states, S1–S6, as previously described (Gong and Peersen, 2010).
