Figure 7. Structures of 3Dpol–VPg complexes.
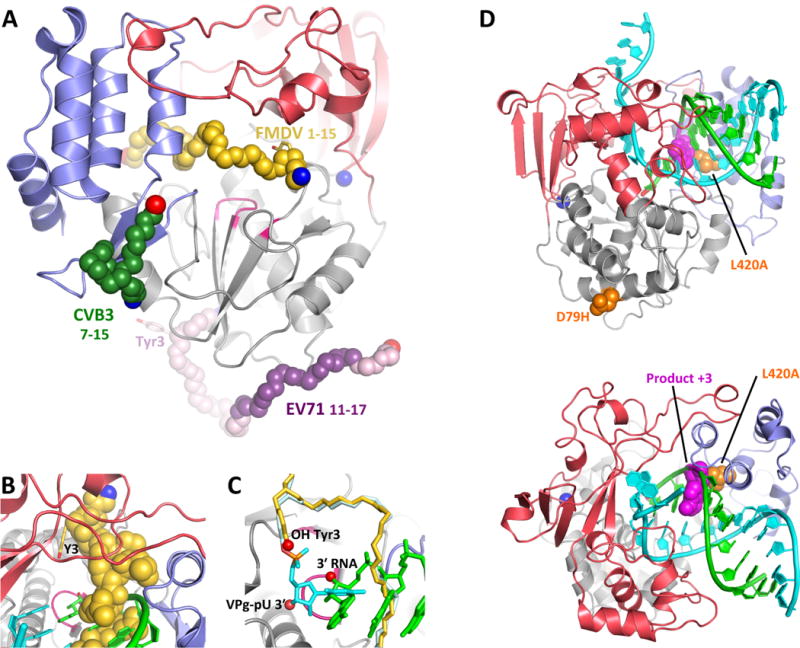
(A) The structure of CV 3Dpol onto which the backbone structures of CV, EV71, and FMDV VPg complexes have been superimposed. The N- and C- ends of the VPg peptides are indicated in blue and red, respectively, and the parts of the EV71 VPg structure in weak electron density are shown in lighter purple. Note that only the FMDV VPg is bound near the active site. (B,C) Detailed views of FMDV VPg (and VPg-pU bounds near the active site, with the product RNA strand from PV elongation complex superimposed for comparison. Note the VPg Tyr3 OH group is bound above and across the active site from the normal RNA 3′ OH group and the uracil base of the VPg-pU complex is perpendicular to the bases of the normal RNA binding orientation. (D) Recombination mutants in PV 3Dpol shown on the wildtype EC structure in two orientations. The D79H mutation is located on the exterior surface of the polymerase far from the RNA binding surface. L420A, on the other hand, makes a direct contact with the product strand RNA at the third nucleotide from the active site (magenta) and biochemical data indicate it reduces recombination by reducing the efficiency of re-initiation on a new template strand.
