Abstract
Purpose
The purpose of this study was to investigate the chromosomal constitution and the developmental potential of intracytoplasmic sperm injection (ICSI) deriving embryos displaying a single pronucleus at the zygote stage.
Methods
Eighty-eight embryos from single pronucleus (1PN) two polar bodies (2PB) ICSI zygotes from 64 preimplantational genetic screening (PGS) cycles (October 2012–December 2014), were retrospectively analyzed. Zygotes were cultured in a time-lapse incubator. Embryo biopsy was performed on day 3 and genetic analysis approached by array comparative genomic hybridization.
Results
Chromosomal analysis revealed that 17% (15/88) of embryos derived from 1PN 2PB zygotes were diagnosed as euploid. After blastomere biopsy at day 3, the blastocyst rate at day 5 was 3.4% (3/88). Only 2.3% (2/88) euploid blastocysts were obtained. In two couples and after counseling and patient agreement, the transfer of a euploid blastocyst from a 1PN 2PB ICSI zygote was performed resulting in the birth of a healthy child.
Conclusions
These results open the possibility to consider embryos coming from 1PN 2PB ICSI zygotes for transfer when no other embryos from 2PN 2PB ICSI zygotes are available and if a PGS diagnosis of euploidy is obtained. Confirmation of biparental inheritance is strongly recommended.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-017-0937-z) contains supplementary material, which is available to authorized users.
Keywords: aCGH, Monopronucleated zygotes, 1PN, PGS, Time-lapse
Introduction
The most important goal of assisted reproduction techniques (ART) is the delivery of a healthy child. In the last years, two technologies, preimplantational genetic screening (PGS) with comprehensive chromosome screening (CCS) and time-lapse embryo monitoring, have been used to investigate parameters that could contribute to assess the implantation potential of embryos.
The application of PGS in embryos from ART cycles aims to improve the implantation rates (raising the birth rate and decreasing the miscarriage rate), avoiding the replacement of aneuploid embryos [1–3]. Furthermore, it has been reported that poor-prognosis patients (advanced maternal age, implantation failure, male factor, recurrent miscarriage) produce embryos with high aneuploidy rates and may especially benefit from this technology [3–5]. Moreover, the optimization of incubators, combining stable culture conditions and high-resolution time-lapse image capturing, allows a continuous monitorized analysis of embryo development until the blastocyst stage. As it has been reported [6–9], the information obtained by these systems may help to select embryos with the highest implantation potential. PGS and time-lapse monitoring used in combination can generate valuable information on the decision of transferring or discarding specific embryos, as both the chromosomal constitution and the dynamics of embryo development can be taken into account [10].
In certain patients, the only embryos available arise from abnormal fertilization, abnormal cleavage and/or blastomere multinucleation. In these situations, there is a general consensus about discarding zygotes showing three pronuclei, and this decision is also adopted for most single pronucleus (1PN)-derived embryos. It has been reported that monopronucleated zygotes obtained from conventional IVF (cIVF) are mainly diploid [11, 12] and can be used for reproductive purposes. Embryos arising from 1PN intracytoplasmic sperm injection (ICSI) zygotes are not usually considered for transfer as there are concerns about their chromosomal constitution and are usually discarded. However, because in certain cases there are no other embryos available, new information would be valuable to help in the decision about transferring or discarding them.
Several articles investigating the possible origin of 1PN zygotes and reporting data about the chromosome constitution of the resulting embryos have been published [13–15]. These reports have shown that certain embryos originating from 1PN two polar bodies (2PB) zygotes could have a normal chromosomal content.
In this work, we have retrospectively analyzed by array comparative genomic hybridization (aCGH) the chromosomal constitution of embryos arising from 1PN 2PB ICSI zygotes obtained from patients submitted to PGS cycles. Although we are aware of the limitations of day 3 biopsy and the use of aCGH for embryo ploidy detection, we have considered that the information gained through the results could be of benefit for the discussion about the possible use of these embryos for reproductive purposes.
Material and methods
PGS cycles performed from October 2012 to December 2014 were retrospectively analyzed. During this period, 385 cycles were accomplished, and 64 cycles meet the criteria of the study: containing at least one embryo where a 1PN and 2PB at the zygote stage were observed, embryos being cultured in time-lapse incubator after ICSI, embryo biopsy done on day 3, and genetic analysis performed by aCGH.
From the 64 PGS cycles analyzed, the mean maternal age was 38.6 years (±3.9) and the PGS indications were as follows: recurrent miscarriage (20.3%), advanced maternal age (25%), male factor (23.4%), implantation failure (26.6%), and previous affected pregnancy (4.7%).
A total of 115 embryos from 1PN 2PB zygotes were recorded, and 88 of them fulfill the criteria for embryo biopsy on day 3 and constituted the study population.
Intracytoplasmic sperm injection was performed 40 h after HCG administration and zygotes were cultured in LifeGlobal total® media (LifeGlobal®) in an EmbryoScope® (Vitrolife) capturing images every 15 min in five focal planes. Continued sequential observation performed by dynamic monitoring allowed excluding sporadic cases of asynchronous 2PN formation or 2PN fusion. Monopronucleated zygotes without extrusion of the 2PB were also excluded from the study. Zygotes were cultured until day 3 when biopsy was performed. Embryos showing five or more blastomeres, and a maximum of 25% of cytoplasmic fragmentation, were biopsied. A single cell per embryo was biopsied. Blastomeres were analyzed by aCGH using 24 sure kit and fluorescent labeling system (Illumina®) according to manufacturer’s protocols. After biopsy, embryos were placed in fresh LifeGlobal total® media (LifeGlobal®) and cultured until day 5 when transfer was performed.
During the period analyzed, the PGS policy of our center was to perform embryo biopsy on day 3 and to select for transfer the embryos reported to be euploid that reached the blastocyst stage on day 5.
The chromosomal status of embryos arising from 1PN 2PB ICSI zygotes was analyzed by aCGH and embryos were classified as euploid, aneuploid, or with non-conclusive results. In the group of aneuploid embryos, two subgroups were established according to the classification described by Johnson et al. [16]: simple aneuploidy (1–2 chromosome abnormalities) or complex aneuploidy (≥3 chromosome abnormalities). Embryos without diagnosis were classified into two subgroups depending on whether there has been DNA amplification but non-conclusive results were obtained or no DNA amplification was observed.
According to morphological analysis and developmental outcome on day 5, embryos were classified as blastocysts (early blastocysts, expanded and hatching blastocysts), morulae, and arrested or degenerated embryos.
Euploid embryos from 2PN 2PB zygotes were always first selected for transfer. Exceptionally, in two cycles from two different patients where no euploid embryos from 2PN zygotes were available, patients were informed about the possibility to transfer one embryo from 1PN 2PB ICSI zygote diagnosed as euploid. This option was extensively discussed with the couples, including the limitations of the diagnosis and the possible risks. Thus, after patient agreement, two single embryo transfers of euploid blastocysts derived from 1PN 2PB ICSI zygotes were performed.
Euploidy and blastocyst rate were calculated and the association between the chromosomal constitution and the developmental stage was analyzed using chi-square test. All tests were bilateral with a significance set to 0.05. The clinical outcome of the cases where embryos coming from 1PN 2PB ICSI zygotes were replaced were analyzed.
The project was approved by the institutional review board of the center.
Results
The mean number of inseminated oocytes in the 64 cycles studied was 14.14 (±4.16), with a mean number of 10 (±3.41) oocytes fertilized, and a mean number of biopsied embryos of 9.75 (±3.02).
The median time of second polar body appearance was 3.53 h post insemination (hpi) (range 1.43–5.13) and the median time of pronucleus appearance was 8.2 hpi (range 5.95–12.63).
The chromosomal analysis of 88 embryos coming from 1PN 2PB ICSI zygotes showed that 17% (15/88) were euploid and 60.2% (53/88) aneuploid. No diagnosis was obtained in the remaining 22.7% embryos (20/88) due to an absence of DNA amplification (13.6%; 12/88) or to non-conclusive results (9.1%; 8/88). According to the aCGH results 28.4% (25/88) showed a simple aneuploidy and 31.8% (28/88) a complex aneuploidy. The presence of a Y chromosome was evidenced in 10.3% of the diagnosed embryos (7/68).
Embryo development analysis showed that 3.4% (3/88) of the biopsied embryos from 1PN 2PB ICSI zygotes reached the blastocyst stage, 36.4% (32/88) arrested at the morula stage and 60.2% arrested their development at earlier stages (53/88).
The relationship between the developmental stage, after blastomere biopsy, and the chromosomal constitution is detailed in Table 1. Of the 15 embryos diagnosed as euploid, only 2 (13.3%) formed a blastocyst whereas none of the aneuploid embryos reached this stage. The majority of aneuploid embryos (69.8%; 37/53) arrested their development at early stages. Among embryos without diagnosis, one reached the blastocyst stage (5%; 1/20) and the remaining ones arrested at the morula stage (35%; 7/20) or at earlier stages (60%; 12/20). In total, only 2.3% (2/88) euploid blastocysts were obtained.
Table 1.
Relationship between developmental stage in day 5, after biopsy on day 3, and chromosomal constitution of embryos derived from 1PN 2PB ICSI zygotes
| Chromosomal constitution | Developmental stage D5 | |||||
|---|---|---|---|---|---|---|
| Blastocyst | Morulae | Arrested | Total | |||
| Euploid | 2 (13.3%) | 9 (60.0%) | 4 (26.7%) | 15 | ||
| Aneuploid | 0 (0.0%) | 16 (30.2%) | 37 (69.8%) | 53 | ||
| Non-diagnosed | Without conclusive diagnosis | 1 (12.5%) | 5 (62.5%) | 2 (25%) | 8 | |
| No amplification | 0 | 2 (16.7%) | 10 (83.3%) | 12 | ||
| TOTAL | 88 | |||||
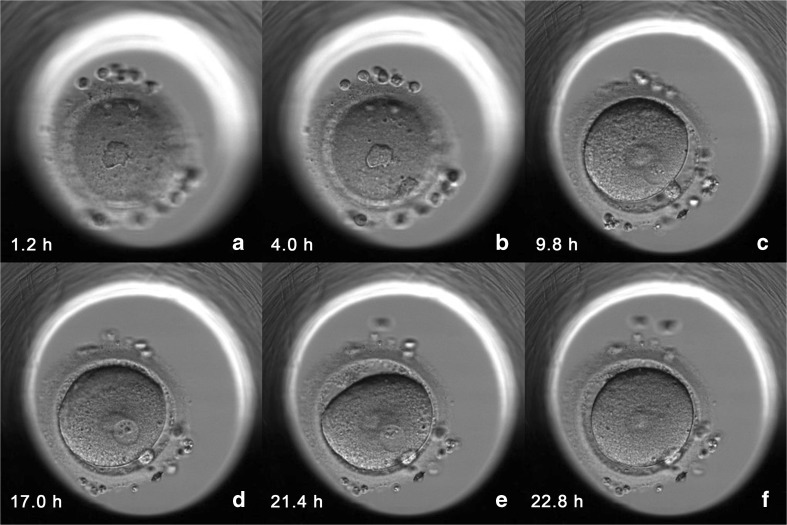
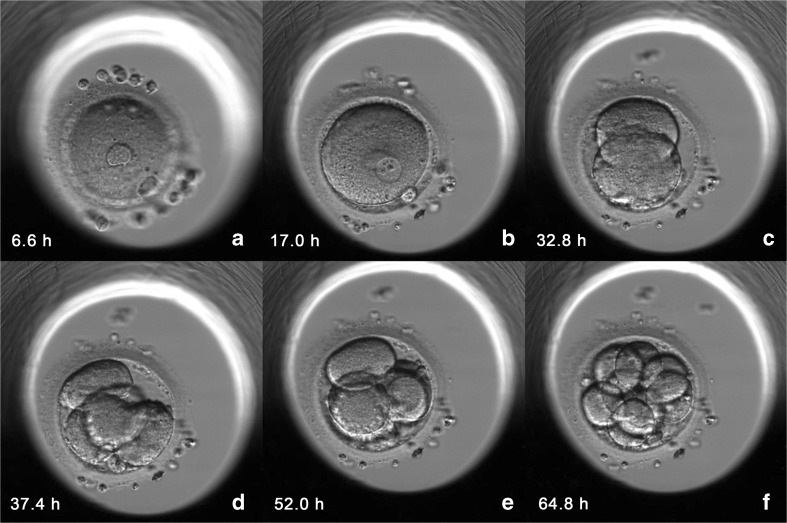
In two patients, all the embryos coming from 2PN zygotes were diagnosed as aneuploid. In one patient, 12 embryos were biopsied with 11 being aneuploid and the only one diagnosed as euploid came from 1PN 2PB ICSI zygote. The other patient had 9 embryos biopsied, with 8 being aneuploid and the remaining embryo, from 1PN 2PB ICSI zygote, diagnosed as euploid. In these two patients, and because no euploid embryos from 2PN 2PB zygotes were available, single embryo transfer of a blastocyst from a 1PN 2PB ICSI zygote diagnosed as euploid was performed on day 5 (Figs. 1 and 2) (Video 1, Online Resource). One pregnancy was achieved resulting in the birth of a healthy child. No prenatal diagnosis was performed. The patient delivered at 40 weeks of gestation a healthy female of 3136 g and 47 cm. Child development has been followed up since birth and, at the moment of preparing this manuscript, the baby was 3 years old and no alterations were observed in her development according to ordinary pediatric check-up and parent’s communication.
Fig. 1.
Time-lapse imaging of second polar body (PB) extrusion and pronuclear (PN) formation and fading of the 1PN 2PB ICSI zygote that produced an ongoing pregnancy and the delivery of a healthy child. The time of image captures (hours post ICSI) is shown at the bottom left of each image. a. First PB. b Extrusion of the second PB. c–e Formation of 1PN. f Fading of PN
Fig. 2.
Time-lapse imaging of in vitro development until day 3 of 1PN 2PB ICSI zygote that produced an ongoing pregnancy and the delivery of a healthy child. The time of image captures (hours post ICSI) is showed at the bottom left of each image. a First and second PB. b 1PN 2PB. c–f 2-, 4-, 5-, and 8-cell embryo
Discussion
The use of dynamic monitoring (time-lapse) during this study allowed a detailed observation of second polar body extrusion and pronuclear formation. Unlike previous studies, where traditional culture systems and observations at specific check points were used [13, 17, 18], our approach unequivocally guarantees that all the zygotes included in the study displayed only a single pronucleus and had extruded the second polar body (Fig. 1). Asynchronous pronuclear formation or pronuclear fusion, as causes for the appearance of a single PN, can be ruled out as PN formation was accurately ascertained through time-lapse imaging and were clearly established as exclusion criteria. No debris or fragments in the perivitelin space were present that could make the observation of PN formation difficult. The presence of 2PB was also clearly established to avoid the inclusion of diploid embryos resulting from the non-extrusion of the second polar body. Timings for second PB extrusion and PN appearance are similar to those observed in 2PN zygotes [19]. According to the images obtained, our hypothesis is that the origin of the monopronucleated zygotes could be due to either a parthenogenetic oocyte activation or to an abnormal formation of the nuclear envelope. This last could result from the assembly of the two genetic materials in a single PN or from the failure to organize a nuclear envelope around one of the parental genomes [20].
The results obtained after blastomere biopsy and aCGH analysis of embryos from 1PN ICSI zygotes reported 22.7% of undiagnosed embryos. This figure is higher than that in the 2PN embryos from our PGS program, which is around 10% (PGS program unpublished data), and is mainly due to the high number of DNA amplification failures in 1PN embryos (13.6%) in comparison to 2PN embryos (5.4%, PGS program unpublished data). We hypothesize that the high percentage of amplification failures observed can be due to DNA fragmentation or degeneration that occurs in embryos from 1PN ICSI zygotes prior to embryo development arrest. This explanation is in harmony with the observation of a high rate of embryo developmental arrest after day 3 in 1PN embryos (60.2%). Furthermore, it can also be presumed that the low ability of biopsied embryos from 1PN ICSI zygotes to reach the blastocyst stage can be attributed to the fact that some of them are probably haploid and, as has been previously described, haploid embryos derived from 1PN zygotes can be negatively selected during in vitro culture [21, 22]. Even so, it cannot be ruled out that biopsy on day 3 may negatively affect the development capability.
The euploidy rate observed in embryos from 1PN 2PB ICSI zygotes (17%) is lower than that reported in early publications (27.9 and 44.4% of diploid embryos) [17, 23]. The discrepancies could be due to differences between the method of pronuclear assessment (time-lapse vs punctual observations), different diagnostic techniques used (aCGH vs fluorescent in situ hybridization (FISH)) and patient population (PGS patients vs IVF patients).
In a previous report, using five-chromosome FISH analysis, no euploid embryos were found among embryos from 1PN 2PB ICSI zygotes [13]. The differences in the euploidy rate observed between the two studies could be attributed to the experimental designs. In Mateo et al. [13], all embryos, irrespective of their developmental stage (arrested or blastocyst) and morphology, were included, and all the cells were analyzed. In the present study, only embryos that fulfill the criteria for embryo biopsy on day 3 (≥5 cells and ≤25% of fragmentation) were analyzed and arrested or fragmented embryos were not considered. As it has been described [24, 25], a high percentage of cytoplasmic fragmentation or developmental arrest is associated with high levels of chromosome abnormalities. Furthermore, the higher euploidy rate observed in the present study compared with the previous one [13], can be due to the underestimation of mosaicism, a common phenomenon in preimplantation embryos [23, 26, 27].
As previously mentioned, it is important to point out that aCGH has inherent limitations for the detection of ploidy alterations [28] and this represents an important limitation of the results obtained. Thus, the euploidy rate reported in our study has to be taken with caution and further studies of 1PN ICSI deriving embryos should be encouraged.
Considering embryo development, biopsied embryos from 1PN 2PB ICSI zygotes showed a blastocyst rate at day 5 of 3.4% which is similar to rates previously reported [13, 29, 30]. However, this percentage is lower than in 2PN embryos also biopsied on day 3 (24.3%, PGS program unpublished data).The low blastocyst rate obtained could probably be due to the haploid status or other chromosomal or genetic abnormalities that hinder the development to the blastocyst stage in 1PN ICSI zygotes. This would be in accordance with the estimation of diploidy rate inferred from the 10.3% of male embryos observed. The expected balanced XY versus XX embryo rate makes it possible to infer that only around 20.6% of the embryos were diploid.
The clinical use of 1PN 2PB zygotes for transfer or cryopreservation is rare as it has been described that most of them are chromosomally abnormal [13, 15]. As a consequence, data about the implantation potential of embryos from 1PN 2PB zygotes are very scarce. Very few pregnancies and births have been reported after the transfer of embryos from 1PN 2PN zygotes obtained after cIVF [11, 29–31]. After ICSI, there is only one publication reporting a livebirth [18]. In all these publications, although accurate observations after insemination were made, time-lapse was not used. Asynchronous pronuclear formation, pronuclear fusion, or even an abnormal timing of PN appearance/fading, which cannot be detected at the usual check points, could not be completely excluded.
In this work, we report two single embryo transfers of euploid blastocysts from 1PN 2PB ICSI zygotes, assessed with time-lapse technology thus ensuring the status of the single pronucleated zygote, resulting in a livebirth of a healthy female in one of them.
Our results indicate that euploid embryos arising from 1PN 2PB ICSI zygotes could be considered for transfer when there are no euploid embryos from 2PN 2PB ICSI zygotes available. Nevertheless, we strongly recommend DNA fingerprinting analysis before the reproductive use of these embryos. Patients have to be widely informed and counseled and sign a specific informed consent form.
It has to be taken into account that the incidence of 1PN 2PB embryos is low (3.1% unpublished data), and as a consequence, a limited number of embryos from 1PN 2PB ICSI zygotes will be potentially usable. Furthermore, extending embryo culture until blastocyst stage would reduce the number of embryos to be analyzed. Moreover, the analysis of trophectoderm cells would allow better diagnosis approaches and avoid unnecessary day 3 biopsies of embryos that could have initiated a degenerative process [28, 32].
In summary, when no euploid 2PN 2PB embryos are available and after appropriate patient counseling, we suggest that embryos coming from 1PN 2PB ICSI zygotes diagnosed as euploid and after DNA fingerprinting analyses, could be considered for reproductive purposes. Furthermore, in order to optimize the cost-benefit of our proposal, we suggest approaching PGS by trophectoderm biopsy.
Finally, we would like to emphasize that the aim of the present paper was not to encourage the transfer of 1PN 2PB ICSI-derived embryos as a general practice but to gain knowledge about their chromosomal constitution and give elements to promote the debate about the possible reproductive use of these embryos when no other ones are available.
chi-square test showed association between both variables (p = 0.012).
Electronic supplementary material
In vitro development until day 3 of 1PN 2PB ICSI zygote that produced an ongoing pregnancy and the delivery of a healthy child (AVI 313780 kb)
Compliance with ethical standards
The project was approved by the institutional review board of the center.
Funding
This work was performed under the auspices of “Càtedra d’Investigació en Obstetrícia i Ginecologia” of the Department of Obstetrics, Gynecology and Reproduction, Dexeus Women’s Health (Barcelona).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-017-0937-z) contains supplementary material, which is available to authorized users.
References
- 1.Yang Z, Liu J, Collins GS, Salem SA, Liu X, Lyle SS, Peck AC, Sills ES, Salem RD. Selection of single blastocysts for fresh transfer via standard morphology assessment alone and with array CGH for good prognosis IVF patients: results from a randomized pilot study. Mol Cytogenet. 2012;5:24. doi: 10.1186/1755-8166-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forman EJ, Hong KH, Ferry KM, Tao X, Taylor D, Levy B, Treff NR, Scott RT. In vitro fertilization with single euploid blastocyst transfer: a randomized controlled trial. Fertil Steril. 2013;100:100–107. doi: 10.1016/j.fertnstert.2013.02.056. [DOI] [PubMed] [Google Scholar]
- 3.Harton GL, Munné S, Surrey M, Grifo J, Kaplan B, McCulloh DH, Griffin D, Wells D. Diminished effect of maternal age on implantation after preimplantation genetic diagnosis with array comparative genomic hybridization. Fertil Steril. 2013;100:1695–1703. doi: 10.1016/j.fertnstert.2013.07.2002. [DOI] [PubMed] [Google Scholar]
- 4.Rubio C, Buendia P, Rodrigo L, Mercader A, Mateu E, Peinado V, Delgado A, Milan M, Mir P, Simon C, et al. Prognostic factors for preimplantation genetic screening in repeated pregnancy loss. Reprod BioMed Online. 2009;18:687–693. doi: 10.1016/S1472-6483(10)60015-6. [DOI] [PubMed] [Google Scholar]
- 5.Rodrigo L, Peinado V, Mateu E, Remohí J, Pellicer A, Simón C, Gil-Salom M, Rubio C. Impact of diferent paterns of sperm chromosomal abnormalities on the chromosomal constitution of preimplantation embryos. Fertil Steril. 2010;94:1380–1386. doi: 10.1016/j.fertnstert.2009.05.061. [DOI] [PubMed] [Google Scholar]
- 6.Lemmen JG, Agerholm I, Ziebe S. Kinetic markers of human embryo quality using time-lapse recordings of IVF/ICSI-fertilized oocytes. Reprod BioMed Online. 2008;17:385–391. doi: 10.1016/S1472-6483(10)60222-2. [DOI] [PubMed] [Google Scholar]
- 7.Meseguer M, Herrero J, Tejera A, Hilligsøe KM, Ramsing NB, Remoh J. The use of morphokinetics as a predictor of embryo implantation. Hum Reprod. 2011;26:2658–2671. doi: 10.1093/humrep/der256. [DOI] [PubMed] [Google Scholar]
- 8.Basile N, Vime P, Florensa M, Aparicio Ruiz B, Garcia Velasco JA, Remohi J, Meseguer M. The use of morphokinetics as a predictor of implantation: a multicentric study to define and validate an algorithm for embryo selection. Hum Reprod. 2015;30:276–283. doi: 10.1093/humrep/deu331. [DOI] [PubMed] [Google Scholar]
- 9.Castello D, Motato Y, Basile N, Remohi J, Espejo-Catena M, Meseguer M. How much have we learned from time-lapse in clinical IVF? Mol Hum Rep. 2016;22:719–727. doi: 10.1093/molehr/gaw056. [DOI] [PubMed] [Google Scholar]
- 10.Yang Z, Zhang J, Salem SA, Liu X, Kuang Y, Salem RD, Liu J. Selection of competent blastocysts for transfer by combining time-lapse monitoring and array CGH testing for patients undergoing preimplantation genetic screening: a prospective study with sibling oocytes. BMC Med Genet. 2014;7:38. doi:10.1186/1755-8794-7-38. [DOI] [PMC free article] [PubMed]
- 11.Staessen C, Janssenswillen C, Devroey P, Van Steirteghem AC. Cytogenetic and morphological observations of single pronucleated human oocytes after in-vitro fertilization. Hum Reprod. 1993;8:221–223. doi: 10.1093/oxfordjournals.humrep.a138026. [DOI] [PubMed] [Google Scholar]
- 12.Munné S, Tang Y, Grifo J, Cohen J. Origin of single pronucleated human zygotes. J Assist Reprod Genet. 1993;10(4):276–279. doi: 10.1007/BF01204942. [DOI] [PubMed] [Google Scholar]
- 13.Mateo S, Parriego M, Boada M, Vidal F, Coroleu B, Veiga A. In vitro development and chromosome constitution of embryos derived from monopronucleated zygotes after intracytoplasmic sperm injection. Fertil Steril. 2013;99:897–902. doi: 10.1016/j.fertnstert.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 14.Azevedo AR, Pinho MJ, Silva J, Sa R, Thorsteinsdottir S, Barros A, Sousa M. Molecular cytogenetics of human single pronucleated zygotes. Reprod Sci. 2014;21:1472–1482. doi: 10.1177/1933719114530185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenbusch B. The chromosomal constitution of embryos arising from monopronuclear oocytes in Programmes of assisted reproduction. Int J Reprod Med. 2014:1–8. [DOI] [PMC free article] [PubMed]
- 16.Johnson DS, Cinnioglu C, Ross R, Filby A, Gemelos G, Hill M, Ryan A, Smotrich D, Rabinowitz M, Murray MJ. Comprehensive analysis of karyotypic mosaicism between trophectoderm and inner cell mass. Mol Hum Reprod. 2010;16:944–949. doi: 10.1093/molehr/gaq062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Staessen C, Van Steirteghem AC. The chromosomal constitution of embryos developing from abnormally fertilized oocytes after intracytoplasmic sperm injection and conventional in-vitro fertilization. Hum Reprod. 1997;12:321–327. doi: 10.1093/humrep/12.2.321. [DOI] [PubMed] [Google Scholar]
- 18.Barak Y, Kogosowski A, Goldman S, Soffer Y, Gonen Y, Tesarik J. Pregnancy and birth after transfer of embryos that developed from single-nucleated zygotes obtained by injection of round spermatids into oocytes. Fertil Steril. 1998;70:67–70. doi: 10.1016/S0015-0282(98)00106-X. [DOI] [PubMed] [Google Scholar]
- 19.Boada M, Gil Y, Mateo S, Joda L, Barri PN, Veiga A. Morphokinetic differences on embryo development after normal or abnormal fertilization. Fert and Ster. 2012;98(3):S159–S160. doi: 10.1016/j.fertnstert.2012.07.588. [DOI] [Google Scholar]
- 20.Van der Heijen GW, Van der Berg IM, Baart EB, Derijck AAHA, Martini E, De Boer P. Parental origin of chromatin in human monopronuclear zygotes revealed by asymmetric histone methylation patterns, differs between IVF and ICSI. Mol Reprod Dev. 2009;76:101–108. doi: 10.1002/mrd.20933. [DOI] [PubMed] [Google Scholar]
- 21.Gras L, Trounson AO. Pregnancy and birth resulting from transfer of a blastocyst observed to have one pronucleus at the time of examination for fertilization. Hum Reprod. 1999;14:1869–1871. doi: 10.1093/humrep/14.7.1869. [DOI] [PubMed] [Google Scholar]
- 22.Liao H, Zhang S, Cheng D, Ouyang Q, Lin G, Gu Y, Lu C, Gong F, Lu G. Cytogenetic analysis of human embryos and embryonic stem cells derived from monopronuclear zygotes. J Assist Reprod Genet. 2009;26:583–589. doi: 10.1007/s10815-009-9355-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim AST, Goh VHH, Su CL, Yu SL. Microscopic assessment of pronuclear embryos is not definitive. Hum Genet. 2000;107:62–68. doi: 10.1007/s004390000335. [DOI] [PubMed] [Google Scholar]
- 24.Munné S. Chromosome abnormalities and their relationship to morphology and development of human embryos. Reprod BioMed Online. 2006;12:234–253. doi: 10.1016/S1472-6483(10)60866-8. [DOI] [PubMed] [Google Scholar]
- 25.Magli MC, Gianaroli L, Ferraretti AP, Lappi M, Ruberti A, Farfalli V. Embryo morphology and development are dependent on the chromosomal complement. Fertil Steril. 2007;87:534–541. doi: 10.1016/j.fertnstert.2006.07.1512. [DOI] [PubMed] [Google Scholar]
- 26.Fragouli E, Alfarawati S, Daphnis DD, Goodall NN, Mania A, Griffiths T, Gordon A, Wells D. Cytogenetic analysis of human blastocysts with the use of FISH, CGH and aCGH: scientific data and technical evaluation. Hum Reprod. 2011;26:480–490. doi: 10.1093/humrep/deq344. [DOI] [PubMed] [Google Scholar]
- 27.Mertzanidou A, Wilton L, Cheng J, Spits C, Vanneste E, Moreau Y, Vermeesch JR, Sermon K. Microarray analysis reveals abnormal chromosomal complements in over 70% of 14 normally developing human embryos. Hum Reprod. 2013;28:256–264. doi: 10.1093/humrep/des362. [DOI] [PubMed] [Google Scholar]
- 28.Gutiérrez-Mateo C, Colls P, Sánchez-García J, Escudero T, Prates R, Ketterson K, Wells D, Munné S. Validation of microarray comparative genomic hybridization for comprehensive chromosome analysis of embryos. Fertil Steril. 2011;95:953–958. doi: 10.1016/j.fertnstert.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 29.Itoi F, Asano Y, Shimizu M, Honnma H, Murata Y. Birth of nine normal healthy babies following transfer of blastocysts derived from human single-pronucleate zygotes. J Assist Reprod Genet. 2015;32:1401–1407. doi: 10.1007/s10815-015-0518-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otsu E, Sato A, Nagaki M, Araki Y, Utsunomiya T. Developmental potential and chromosomal constitution of embryos derived from larger single pronuclei of human zygotes used in in vitro fertilization. Fertil Steril. 2004;81:723–724. doi: 10.1016/j.fertnstert.2003.07.042. [DOI] [PubMed] [Google Scholar]
- 31.Dasig D, Lyon J, Behr B, Milki A. Monozygotic twin birth after the transfer of a cleavage stage embryo resulting from a single pronucleated oocyte. J Assist Reprod Genet. 2004;21:427–429. doi: 10.1007/s10815-004-8758-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin BL, Hao HY, Zhang YN, Wei D, Zhang CL. Good quality blastocyst from non-/mono-pronuclear zygote may be used for transfer during IVF. Syst Biol Reprod Med. 2016;62:139–145. doi: 10.3109/19396368.2015.1137993. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In vitro development until day 3 of 1PN 2PB ICSI zygote that produced an ongoing pregnancy and the delivery of a healthy child (AVI 313780 kb)