Figure 2.
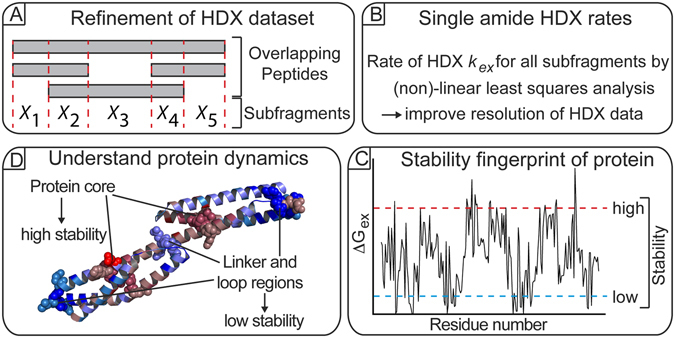
Workflow of HR-HDXMS. (A) Amino acid subfragments are determined from overlapping peptides generated by proteolytic digest of the protein. (B) The rate of hydrogen-deuterium exchange (k ex) of each subfragment is calculated by linear and non-linear least squares analysis. (C) The free energy value ΔGex derived from k ex is used to generate a stability fingerprint of the protein. (D) The protein fingerprint helps in understanding protein dynamics and allows predictions about residues that might be essential for protein stability and/or function.
