Figure 3.
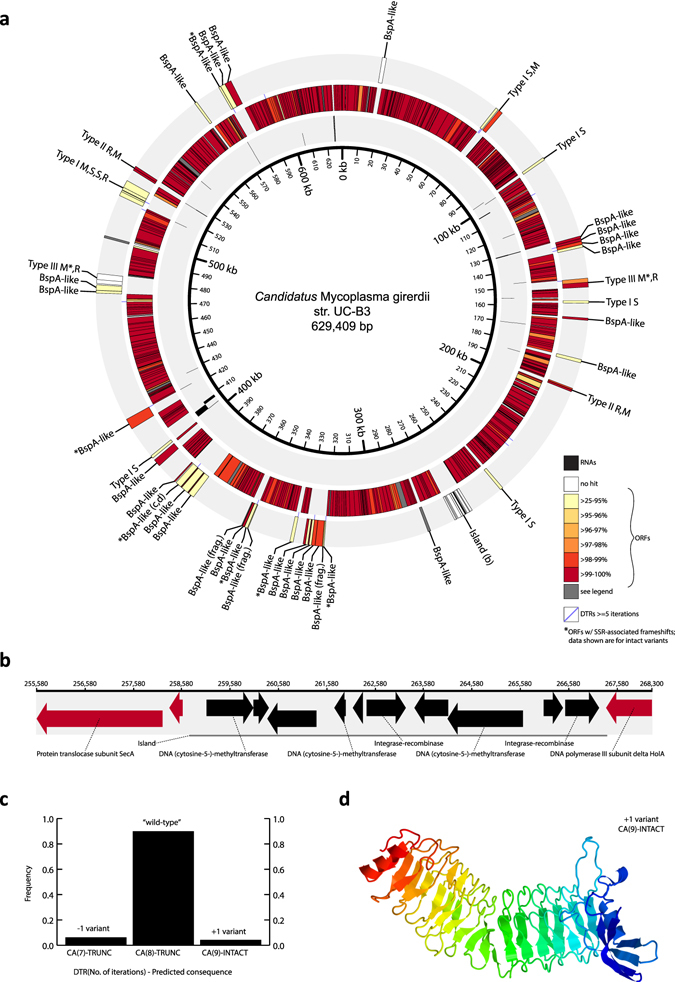
Genomic features distinguishing Ca. M. girerdii strain UC-B3 from strain VCU-M1 include those with evidence of phase variation within population UC-B3. (a) Circular genome map of Ca. M. girerdii strain UC-B3. Inner track shows RNA genes (black; staggered for clarity). Center and outer tracks show open reading frames (ORFs; color graded on percent amino acid identity to reciprocal best hit in VCU-M1). ORFs in gray have a close nucleotide hit in strain VCU-M1 but were not predicted there by the authors. Purple lines, located between center and outer tracks, represent dinucleotide tandem repeats (DTRs) of five or more iterations (Supplementary Table S6). ORFs from variable classes were manually placed in the outer track. Those with asterisks display evidence of phase variation (Supplementary Table S7); those without labels (n = 5) encode fructose-bisphosphate aldolase; and those composing restriction modification systems are labeled clockwise as follows: R, restriction endonuclease; M, modification methylase; S, specificity subunit. The subjects of panels b–d are labeled in bold. SSR, simple sequence repeat. (b) Linear map detailing genomic island in UC-B3, which is absent from VCU-M1. ORFs drawn as arrows; those without labels encode hypothetical proteins. (c) Evidence of phase variation for selected feature no. 11 in Supplementary Table S7. Plotted are length variant frequencies for DTR ‘CA’ associated with BspA-like protein labeled in bold in panel a. Homologous ORF in VCU-M1 contains CA(10) (Supplementary Table S7). TRUNC, frameshift-induced premature stop codon resulting in truncated BspA-like protein; INTACT, likely produces intact BspA-like protein shown in panel (d). The intact protein was modeled against the highest scoring template (pneumococcal vaccine antigen PcpA) using Phyre255. Left/red, N-terminus; right/blue, C-terminus.
