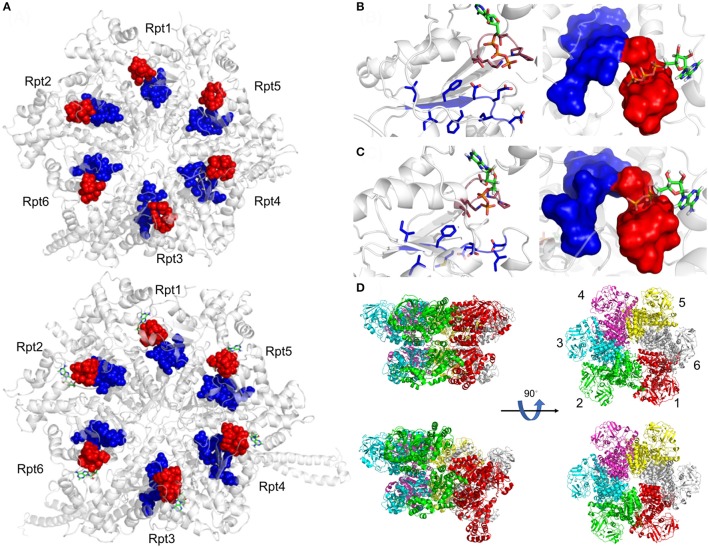
Figure 3.
Active site organization of AAA-ATPase rings. (A) Bottom view of the proteasomal AAA-ATPase rings from yeast (upper panel) and human (lower panel). The Walker domain A is highlighted by red spheres and B by blue spheres. Magnified views of the Walker domains are shown for human AAA-ATPase bound to either ATP (B) or ADP (C) in two orientations. (D) Dynamics of Valosin-containing protein-like ATPase of Thermoplasma acidophilum (VAT) are visualized by conformational switches between the stacked and spiral (split-) ring versions. Side and top views of the AAA-ATPase subunit colored in red show movements out of the plane upon ATP hydrolysis aiding substrate translocation into the proteasome through its central pore. The split ring form (bottom left) undergoes a conformational change back into the stacked ring (top left), when ADP dissociates from the subunit and ATP binds back to allow the next round of hydrolysis. This figure was prepared using the PDB IDs: 4CR2, 5L4G, 5G4G, and 5G4F through PyMOL (Ver. 1.8.0.2) molecular graphics software (Schrodinger, LLC, New York).