Abstract
Background and objectives
Kidney transplantation is the preferred treatment for ESRD, and donor kidney shortage urges proper donor-recipient matching. Zero-hour biopsies provide predictive values for short- and long-term transplantation outcomes, but are invasive and may not reflect the entire organ. Alternative, more representative methods to predict transplantation outcome are required. We hypothesized that proteins accumulating in preservation fluid during cold ischemic storage can serve as biomarkers to predict post-transplantation graft function.
Design, setting, participants, & measurements
Levels of 158 proteins were measured in preservation fluids from kidneys donated after circulatory death (Maastricht category III) collected in two Dutch centers (University Medical Center Utrecht and Erasmus Medical Center Rotterdam) between 2013 and 2015. Five candidate biomarkers identified in a discovery set of eight kidneys with immediate function (IF) versus eight with delayed graft function (DGF) were subsequently analyzed in a verification set of 40 additional preservation fluids to establish a prediction model.
Results
Variables tested for their contribution to a prediction model included five proteins (leptin, periostin, GM-CSF, plasminogen activator inhibitor-1, and osteopontin) and two clinical parameters (recipient body mass index [BMI] and dialysis duration) that distinguished between IF and DGF in the discovery set. Stepwise multivariable logistic regression provided a prediction model on the basis of leptin and GM-CSF. Receiver operating characteristic analysis showed an area under the curve (AUC) of 0.87, and addition of recipient BMI generated a model with an AUC of 0.89, outperforming the Kidney Donor Risk Index and the DGF risk calculator, showing AUCs of 0.55 and 0.59, respectively.
Conclusions
We demonstrate that donor kidney preservation fluid harbors biomarkers that, together with information on recipient BMI, predict short-term post-transplantation kidney function. Our approach is safe, easy, and performs better than current prediction algorithms, which are only on the basis of clinical parameters.
Keywords: renal transplantation, transplant outcomes, ischemia-reperfusion, cytokines
Introduction
Kidney transplantation is the preferred treatment for patients with ESRD. It is expected that the need for donor organs will increase in the coming years as life expectancy increases in the Western world, with a concomitant rise in age-related renal disease. The availability of donor organs is lagging, and a partial solution to bridge this gap is the increased use of extended criteria donor (ECD) organs and improved pre- and post-transplantation care to reduce loss of transplanted grafts (1). However, delayed graft function (DGF) is much more prevalent in patients receiving such organs, and is associated with high costs and poorer long-term graft function and survival (2,3). To optimize donor potential, it is important to predict which donor kidneys will function directly (immediate function; IF), and which lead to DGF, or even graft failure. This will improve patient-tailored care, and allow rational decisions in acceptance and allocation of donor kidneys. Especially in ECD kidneys, predictive information on organ function could drastically increase transplant availability (4).
Many methods to predict DGF on the basis of biomarkers or clinical parameters have been explored. In 2010, the DGF risk score, predicting DGF on the basis of donor and recipient parameters, was published (5). Zero-hour biopsy specimens are a rich source for morphologic and molecular information to predict short- and long-term kidney function (6–8). However, the information obtained from biopsy specimens is inherently limited due to the focal nature: The procedure carries a risk for damaging the donor kidney, and implementation is under debate (9). Alternative, noninvasive sources for biomarkers are preservation fluid, blood, and urine. Nevertheless, despite the identification of several markers associated with transplant outcome (7,8,10–17), no routine biomarker testing is currently performed.
Here, we explored whether donor kidney preservation fluid, which can be easily obtained through a noninvasive procedure, harbors biomarkers for DGF. We hypothesized that proteins (cytokines, growth factors) secreted into transplant kidney preservation fluid by the donor kidney reflect its condition, and may therefore serve as biomarkers to assess donor kidney quality and predict post-transplantation function.
Cytokines and growth factors are important to maintain homeostasis by mediating communication between different cells and organs. ILs, for example, are secreted during infection and inflammation, eliciting immune responses (18,19), and hypoxia induces growth factor release (20). Donor kidneys, including donation after circulatory death (DCD) kidneys, are perfused with cold preservation fluid and stored on ice. Preservation fluid, which is in direct contact with the kidney’s vasculature during this entire period, accumulates such secreted proteins.
In this study, we interrogated the protein content of organ preservation fluid aiming to identify biomarkers and establish a prediction model for DGF.
Materials and Methods
Study Design, Sample Collection, and Processing
Sample and data collection were approved by local ethical committees and in agreement with the Declaration of Helsinki. All recipients provided informed consent to retrieve data from medical records for The Netherlands Organ Transplantation Registry. Samples were prospectively collected from DCD-derived donor kidneys (Maastricht category III) transplanted between January of 2013 and July of 2015 in the University Medical Center Utrecht (Utrecht, The Netherlands; n=37), and between November of 2014 and August of 2015 in the Erasmus University Medical Center (Rotterdam, The Netherlands; n=19). After cold static preservation, kidneys were inspected in the hospital for transplantation and intravascular preservation fluid was harvested by perfusion of 40–60 ml of preservation fluid (University of Wisconsin solution) or physiologic salt solution with 20 U/ml heparin into the renal artery. Preservation fluid, flowing out from the renal vein (1.5–27 ml), was collected sterile. Cells and platelets were removed by centrifugation (2000 × g for 30 minutes or 4000 × g for 15 minutes, depending on center), and the cell-free supernatant was aliquoted and stored at −20°C or −80°C until further analysis.
For initial analysis (discovery), we selected eight samples from kidneys with IF and matched these on the basis of donor age and cold ischemic period with eight samples from kidneys with DGF, defined as “need for dialysis indicated by poor kidney function (low urine output, high serum creatinine) within the first week after transplantation.” After sample collection for the discovery set was complete, we continued sample collection to establish a cohort for verification and subsequent definition of a DGF prediction model. Sample size calculation for the verification cohort was on the basis of the method by Hsieh et al. (21) for logistic regression as employed in the powerMediator package in R. For calculation, we used a baseline DGF risk of 65%, the lowest odds ratio among factors identified in the first discovery cohort (GM-CSF, odds ratio, 4.0), α of 0.01, and a β of 95%, to provide for multiple hypotheses, indicating that 40 samples were required which were collected at both participating centers. In a low immunologic risk situation, no induction was given and immunosuppression in general consists of tacrolimus and mycophenolate. Steroids were tapered to zero if no problems arose. In intermediate immunologic risk, induction was performed with basiliximab, and immunosuppression in general consists of tacrolimus, mycophenolate, and steroids. In high immunologic risk situations (very high panel-reactive antibodies), alemtuzumab is used as an induction agent.
Donor and Recipient Data
For calculations using prediction algorithms, donor and organ data were obtained from the Eurotransplant database, and recipient data were retrieved from The Netherlands Organ Transplant Registry. Apart from cold ischemia time, donor age, and outcome, no information was available until cytokine analyses were performed. Kidney Donor Risk Index (KDRI), which is on the basis of donor age, height, weight, race, hypertension, diabetes, cause of death, serum creatinine, hepatitus C virus status, and donor type (here: DCD), was calculated using the online KDRI calculator (https://optn.transplant.hrsa.gov/resources/allocation-calculators/kdpi-calculator/). The DGF risk calculator (https://www.transplantcalculator.com/Transplant-Calculators/Delayed-Graft-Function.aspx), on the basis of recipient (panel-reactive antibodies, dialysis duration, body mass index (BMI), race, sex, age, previous [extrarenal] transplants, diabetes, and pretransplant blood transfusion), donor (terminal creatinine, age, weight, donor type, hypertension status, and cause of death [stroke/anoxia]), and organ (HLA mismatches, cold ischemia time, warm ischemia time, and machine perfusion) parameters, was used to calculate the DGF risk score (5).
Multiplex Immunoassay
Measurements were performed using an in-house developed and validated multiplex immunoassay on the basis of Luminex technology (xMAP; Luminex, Austin, TX). The assay was performed as described (22,23). Briefly, samples were thawed (without any previous freeze-thaw cycle) and incubated with antibody-conjugated MagPlex microspheres for 1 hour at room temperature while shaking, followed by 1 hour of incubation with biotinylated antibodies, and 10 minutes of incubation with phycoerythrin-conjugated streptavidin diluted in high-performance ELISA buffer (Sanquin, The Netherlands). Each cytokine assay contained a standard curve on the basis of custom-made control peptides and controls for heterophilic antibodies. Acquisition was performed with the Biorad FlexMAP3D (Biorad, Hercules) using xPONENT software version 4.2 (Luminex). Data were analyzed by 5-parametric curve fitting using Bio-Plex Manager software, version 6.1.1 (Biorad). All samples were analyzed before matching to recipient characteristics.
For the discovery phase, samples were studied for the presence and abundance of 158 secreted proteins. Proteins significantly different between IF and DGF groups were selected for subsequent Luminex analysis in a verification cohort of 40 additional samples (UMC Utrecht: n=21; Erasmus Medical Center: n=19).
Statistical Analyses
For analysis of Luminex data, values below the standard curve that could not be extrapolated were set to zero. Values out of range above the standard curve were set to the highest value in the standard curve.
Cluster analysis was performed as described (24). Briefly, measured values were normalized within a 0–1 range. The distance measure used to quantify the distance between two data points was (1−rho), where rho is the pair-wise complete Pearson correlation coefficient between patients. The Ward (minimum variance) method was used for hierarchic clustering of donor samples. Heat maps were created with colors corresponding to the relative cytokine abundances.
Prediction models on the basis of raw measured values were made using multivariable logistic regression, with back- and forward stepwise factor inclusion. Different models were compared with a chi-squared test; model discriminative potential was assessed by receiver operating characteristic (ROC) curves. Robustness of ROC curves to patient population variations was assessed by internal resampling with replacement (bootstrapping, 10,000 iterations). P values <0.05 were considered statistically significant. Analyses were conducted using Microsoft Excel, SPSS statistics 21, and R (v.3.1.2).
Results
Donor, Recipient, and Graft Characteristics
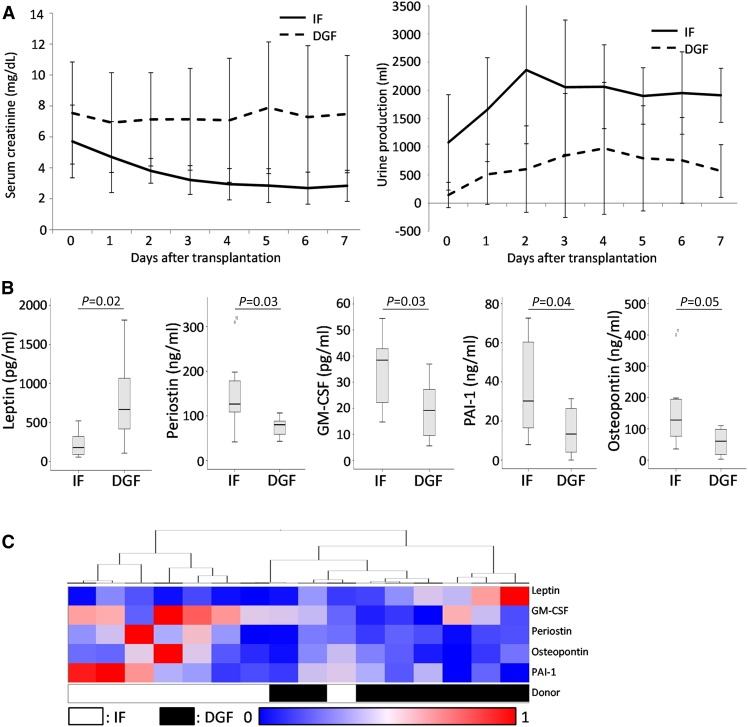
Donor, recipient, and graft characteristics of the discovery panel are presented in Table 1. In the DGF group, serum creatinine levels remained high during the first week after transplantation, whereas the expected reduction is observed in the IF group. Accordingly, urine production increased in the IF group and did not in the DGF group (Figure 1A). Patients in the DGF group underwent a median of two (range, 1–8) dialysis sessions after transplantation.
Table 1.
Donor, recipient, and organ characteristics discovery panel
| Parameter | IF (n=8) | DGF (n=8) | P Value |
|---|---|---|---|
| Donor | |||
| Age, yr | 54.0 [49.5–65.3] | 61.5 [54.5–63.3] | 0.89 |
| BMI, kg/m2 | 25.9 (2.2) | 26.3 (4.2) | 0.83 |
| Hypertension, % | 37.5 (n=3) | 12.5 (n=1) | 0.28 |
| Diabetes, % | 12.5 (n=1) | 12.5 (n=1) | >0.99 |
| Creatinine, µM | 80.1 (30.2) | 54.8 (17.9) | 0.06 |
| KDRI | 1.34 (0.42) | 1.32 (0.27) | 0.93 |
| Cause of death: anoxia, % | 37.5 (n=3) | 25 (n=2) | 0.62 |
| Recipient | |||
| Age, yr | 55.5 [50.3–63.3] | 65.5 [57.0–68.3] | 0.43 |
| BMI, kg/m2 | 24.7 (2.7) | 24.8 (3.5) | 0.96 |
| Men, % | 75 (n=6) | 62.5 (n=5) | 0.62 |
| Peak PRA, % | 0 (0) | 0 [0–12.3] | 0.10 |
| Dialysis duration, mo | 24 [18.5–27.5] | 25 [26.3–70] | 0.12 |
| Prior transplants, % | 0 (n=0) | 12.5 (n=1) | 0.33 |
| Dialysis type: HD, % | 37.5 (n=3) | 75 (n=6) | 0.15 |
| Diabetes, % | 12.5 (n=1) | 25 (n=2) | 0.55 |
| Organ | |||
| HLA mismatches | 4 [2.8–4.3] | 3 [1.8–3] | 0.20 |
| CIT, h | 13.5 [12.9–15.0] | 15.8 [11.0–18.1] | 0.79 |
| WIT, min | 30.0 [29.0–34.5] | 29.0 [24.0–30.5] | 0.25 |
Clinical parameters relevant for calculation of KDRI and DGF risk score of the samples of the discovery panel. SDs between brackets; interquartile ranges between square brackets. IF, immediate function; DGF, delayed graft function; BMI, body mass index; KDRI, kidney donor risk index; PRA, panel-reactive antibodies; HD, hemodialysis; CIT, cold ischemia time; WIT, warm ischemia time.
Figure 1.
Analysis of secreted proteins in the discovery panel. (A) Serum creatinine (left) and urine production (right) evolution of the immediate function (IF) and delayed graft function (DGF) discovery sets presented as average±SD. (B) Boxplots representing levels of proteins significantly different between IF and DGF groups in the discovery panel. (C) Heatmap representing clustering of samples from IF and DGF donor kidneys.
Identification of Differentially Abundant Proteins
Of the 158 secreted proteins measured in the discovery panel, three proteins were not detected in any sample, and 26 proteins were detected in fewer than 12 samples. For eight proteins, values were above the standard curve and could not be extrapolated in more than four samples. Undetected proteins and out-of-range values were evenly distributed between groups (Supplemental Table 1).
Five proteins were differentially abundant between DGF and IF groups: leptin, periostin, GM-CSF, plasminogen activator inhibitor-1 (PAI-1), and osteopontin (Figure 1B). Five markers that have previously been associated with DGF in blood and urine, kidney injury molecule-1 (KIM-1), IL-18, YKL-40, cystatin C, and neutrophil gelatinase-associated lipocalin (NGAL), did not differ between groups (Supplemental Figure 1).
To investigate whether these proteins are potential biomarkers for DGF, unsupervised hierarchic clustering of the significantly different proteins was performed, showing that on the basis of protein profiles, most donor samples cluster together, with six correctly clustered individuals for IF group and all in the DGF group (Figure 1C).
Verification of Candidate Biomarkers
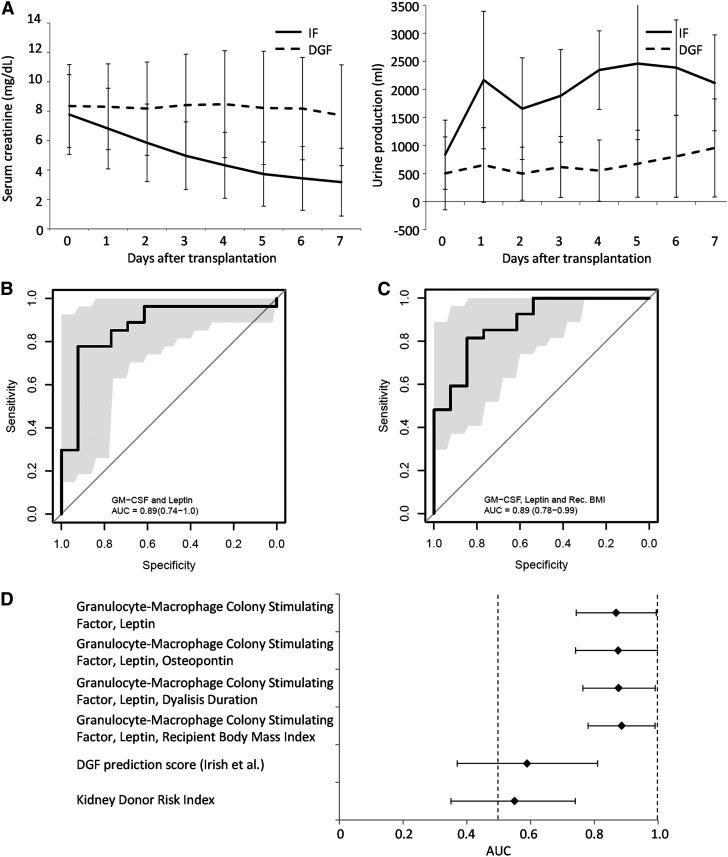
A combined panel of five proteins that were significantly different between IF and DGF groups was used for subsequent Luminex analysis in a verification cohort of 40 additional samples (Table 2). Two patients underwent one dialysis session indicated by hyperkalemia, and are classified as IF; three patients had volume overload or hyperkalemia combined with poor kidney function and were classified as DGF; they all received multiple dialysis sessions. Similar to the discovery set, serum creatinine levels remained high and urine production remained low in the first week after transplantation in the DGF group, and reducing serum creatinine and increasing urine production were observed in the IF group (Figure 2A). Patients in the DGF group underwent 1 to >36 dialysis sessions (median 3) after transplantation. The incidence of DGF in the verification set is 67.5%, in line with the reported incidence of DGF of 63.4% in controlled DCD kidney recipients in The Netherlands (25). At 12 months, overall survival in the IF group was 92.3%, and 81.5% in the DGF group.
Table 2.
Donor, recipient, and organ characteristics verification panel
| Parameter | IF (n=13) | DGF (n=27) | P Value |
|---|---|---|---|
| Donor | |||
| Age, yr | 56.0 [50.0–59.0] | 58.0 [50.8–62.3] | 0.41 |
| BMI, kg/m2 | 25.1 (4.6) | 27.0 (4.4) | 0.20 |
| Hypertension, % | 15.4 (n=2) | 33.3 (n=9) | 0.17 |
| Diabetes, % | 0 (n=0) | 7.4 (n=2) | 0.33 |
| Creatinine, µM | 62.2 (23.5) | 63.1 (21.8) | 0.98 |
| KDRI | 1.22 (0.38) | 1.32 (0.30) | 0.39 |
| Cause of death: anoxia, % | 61.5 (n=8) | 44.4 (n=12) | 0.32 |
| Recipient | |||
| Age, yr | 63.0 [51–68] | 65.5 [52.3–72.0] | 0.77 |
| BMI, kg/m2 | 25.6 (4.3) | 28.3 (4.5) | 0.09 |
| Men, % | 69.2 (n=9) | 63.0 (n=17) | 0.71 |
| Peak PRA, % | 7.3 (15.0) | 6.1 (10.2) | 0.76 |
| Dialysis duration, mo | 13.0 [2.7–18.0] | 31.5 [24.25–51.25] | 0.01 |
| Prior transplants, % | 15.4 (n=2) | 18.5 (n=5) | 0.81 |
| Dialysis type: HD, % | 84.6 (n=11) | 77.8 (n=21) | 0.62 |
| Diabetes, % | 23.1 (n=3) | 25.3 (n=7) | 0.85 |
| Organ | |||
| HLA mismatches | 4 [3–4] | 3.5 [3–4] | 0.37 |
| CIT, h | 11.1 [9.8–14.7] | 13.6 [10.8–17.2] | 0.56 |
| WIT, min | 25.0 [20.0–28.0] | 26.6 [21.8–31.5] | 0.55 |
Clinical parameters relevant for calculation of KDRI and DGF risk score of the 40 samples used for verification. SDs between brackets; interquartile ranges between square brackets. IF, immediate function; DGF, delayed graft function; BMI, body mass index; KDRI, kidney donor risk index; PRA, panel-reactive antibodies; HD, hemodialysis; CIT, cold ischemia time; WIT, warm ischemia time.
Figure 2.
Receiver operating characteristic curves of prediction models. (A) Serum creatinine (left) and urine production (right) evolution of immediate function (IF) and delayed graft function (DGF) verification sets presented as average±SD. Receiver operating characteristic curves representing prediction models on the basis of (B) GM-CSF and leptin, and (C) GM-CSF, leptin, and recipient body mass index, with 95% confidence intervals as determined by bootstrap analysis in gray. (D) Area under the curve (AUC) and 95% confidence intervals of prediction models on the basis of various combinations of factors (indicated); shaded areas indicate bootstrap 95% confidence intervals at all given sensitivity/specificity thresholds.
For all assayed proteins (Supplemental Table 2) no more than three values were under the quantification threshold and set to zero. Stepwise multivariable logistic regression (Supplemental Table 3) was performed to generate a multivariable model, identifying GM-CSF and leptin to provide the best predictive value, demonstrated by an ROC curve with an area under the curve (AUC) of 0.87 (P<0.001) (Figure 2B). Addition of osteopontin slightly, but not significantly, improved the model, shown by an AUC of 0.88 (P<0.001) (Figure 2D, Supplemental Figure 2A).
Biomarkers in Combination with Clinical Parameters
Donor and recipient clinical characteristics show that recipient BMI and dialysis duration are (nearly) significantly different between the IF and DGF groups in the verification cohort (and the complete patient set, Supplemental Table 4), with P values of 0.07 and <0.01, respectively. Addition of dialysis duration as a third variable increased the predictive power slightly, but not significantly (AUC=0.88; Figure 2D, Supplemental Figure 2B). Recipient BMI significantly increased the predictive power, resulting in an AUC of 0.89 (Figure 2C).
In our cohort, all models, whether on the basis of secreted proteins alone, or on a combination of secreted proteins and recipient characteristics, perform significantly better than the currently available algorithms solely on the basis of clinical parameters. This is illustrated for the model described by Irish et al. (5) and for the KDRI (26), which show AUCs of 0.59 and 0.55, respectively, both not significantly different from random chance (Figure 2D, Supplemental Figure 2, C and D).
Discussion
This is the first study to systematically investigate the potential of kidney transplant preservation fluid as a source for biomarkers to predict DGF. We demonstrate that proteins in donor kidney preservation fluid can be used as noninvasive biomarkers in a model to predict DGF. Using a combination of biomarkers and recipient characteristics, we constructed models that predict the occurrence of DGF with high accuracy.
Several biomarkers have been previously associated with DGF, with varying predictive accuracy depending on the compartment (blood, urine, or preservation fluid) under investigation. NGAL and IL-18 are the two most commonly reported biomarkers for kidney injury and post-transplantation graft function. Urinary NGAL has been demonstrated to predict DGF with reasonable accuracy (AUC approximately 0.8) (27), and also plasma and serum NGAL levels at day 1 after transplantation are higher in recipients that develop DGF (16,17,28). An association between DGF and NGAL levels in donor kidney perfusate has been described for machine-perfused kidneys (29). However, similar to our results, no association could be observed in fluid from cold, statically preserved donor kidneys (17). In line with this observation, NGAL and KIM-1 levels in preservation fluid of cold statically preserved donor kidneys also did not show an association with DGF in our study. In zero-hour biopsy specimens this marker did show an association with DGF (11), although its predictive value has been questioned (27,30).
Urinary IL-18, however, has repeatedly been described as an accurate biomarker for post-transplantation kidney function, especially in combination with additional biomarkers such as NGAL and serum creatinine (27,28). Recently, Parikh et al. demonstrated an association between DGF and several markers in cold machine-perfused donor kidney perfusates, including KIM-1, IL-18, and liver fatty acid binding protein (L-FABP) (29), none of which associated with DGF in our discovery panel. Similarly, YKL-40 (in serum and urine) could be detected, but showed no association with DGF. The fact that none of these biomarkers associated with DGF in our study can be attributed to the source of these biomarkers. It is clear that donor kidney preservation fluid is different (in time and source) from blood and urine samples. Furthermore, preservation fluid from machine-perfused kidneys appears to have a different biomarker profile compared with static cold-preserved kidneys. Additionally, biomarkers identified in serum or urine after transplantation rather reflect a response of the recipient to the donor kidney (or vice versa) than the condition of the donor kidney at time of transplantation.
Besides biomarkers, several algorithms to predict post-transplantation graft function on the basis of clinical parameters have been described. The DGF risk calculator uses 22 donor, recipient, and organ parameters to calculate the relative risk at DGF (5). This model has been verified and validated in various external cohorts (31,32). However, as stated in the original paper, this algorithm holds true for (large) patient populations, but has limited value for individual patients. A well established algorithm to assess long-term kidney function after transplantation is the KDRI (26). This algorithm, on the basis of 11 parameters, does not predict short-term kidney function per se, but given the correlation between DGF and poorer prognosis for long-term kidney function, a correlation with DGF may be expected. We could not identify a correlation between KDRI and DGF, fueling the current debate on the association between DGF and long-term survival (2,3,33). In this respect, it should be noted that studies on the association between DGF and long-term survival rates specifically in DCD recipients involve relatively small patient populations, and in the survival curves generally a steep decline can be observed for DCD kidney recipients with DGF within the first 6 months, which is absent in patients with IF. A more gradual decrease in survival of the IF groups renders both groups equal at the 6-year time point (33,34). Similarly, in our cohort, kidney function at 12 months after transplantation does not differ between groups, in line with the recent report that no differences in interstitial fibrosis could be detected between these groups (35). Overall survival at 12 months was higher in the IF group, underscoring the relevance of early identification of DGF allowing for early interventions such as remote preconditioning or ex vivo treatment of the donor kidney, preferably during machine perfusion, which is currently not yet fully implemented in The Netherlands. It has been proposed that for individualized prediction of post-transplantation kidney function, reliable biomarkers in combination with clinical data are essential (36). In our cohort, recipient BMI significantly contributed to the prediction model, in line with the findings by Irish et al. (5) and the association of body weight with DGF in a retrospective study (37,31).
We identified two proteins as important factors: GM-CSF is higher in the IF group, whereas leptin is higher in the DGF group. Although our results do not allow mechanistic conclusions with respect to the pathophysiology of DGF, our data may suggest a role for recruitment of monocytes and macrophages in the early engraftment and short-term function of the donor kidney after transplantation. GM-CSF induces osteopontin (which was indeed found upregulated in the IF group) expression and thereby promotes cell survival (38).
Furthermore, GM-CSF can stimulate tissue regeneration by promoting angiogenesis through activating VEGF-mediated angiopoietin-Tie signaling (39). A synergetic role for GM-CSF and osteopontin in the recruitment of macrophages and monocytes to the implanted kidney, which promotes remodeling and better engraftment, is conceivable. A possible mechanism skewing infiltrated monocytes to M2 (regenerative) macrophages to promote IF is supported by the lower leptin levels in this group, as, besides an “energy indicator,” leptin has been reported to promote a proinflammatory environment and skew macrophages toward the inflammatory M1 phenotype, either directly or through mast cells (40–43).
We have conducted an extensive screen investigating 158 potential biomarkers for DGF. Although extensive, the screen was not exhaustive, and unbiased methods such as proteomics may identify additional biomarkers. Because we chose not to correct for multiple testing in the exploratory stage (discovery) and proceeded with the stringent confirmative phase of the study, we deliberately took the risk of including false positives. Also, we realize that the risk for false negatives is high, and we may have missed (better) candidate biomarkers for verification, and subsequently have not identified the potential most optimal model factors here. Importantly, our study included only 56 participants, transplanted in two medical centers; the findings of this study should be validated in external, ideally prospective, cohorts.
Proteins were selected on the basis of the availability of reliable detection antibodies that would allow fast and straightforward translation to clinical implementation. Sample collection is noninvasive and, in contrast with zero-hour biopsies, without major ethical concerns. Evaluation of these protein biomarkers can be easily achieved in the time between arrival at the hospital and transplantation using an ELISA- or Luminex-based assay, which take about 1 hour from sample delivery to readout, and do not require a pathologist for interpretation (44). Importantly, an additional objective test could help in decisions on acceptance and allocation of donor kidneys, thereby increasing donor potential. Discarding kidneys for transplantation cannot be justified on the basis of such a test, although acceptance may be increased for ECD kidneys with a kidney donor profile index <85 and standard criteria donor kidneys with a kidney donor profile index >85, for which discard rates could be reduced (4).
In conclusion, we describe a noninvasive, easy-to-implement method for the prediction of short-term kidney function after transplantation, on the basis of levels of GM-CSF and leptin in donor kidney preservation fluid and recipient BMI.
Disclosures
None.
Supplementary Material
Acknowledgments
The authors would like to thank Yu-Xuan Chen for excellent assistance, and all transplant surgeons at the University Medical Center Utrecht, Utrecht, The Netherlands, and Erasmus Medical Center, University Medical Center, Rotterdam, The Netherlands, involved in the collection of preservation fluids.
This research was made possible by an unrestricted research grant from Shire (AR 2014-118) to B.W.M.v.B.
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
See related editorial, “Can Preservation Fluid Biomarkers Predict Delayed Graft Function in Transplanted Kidneys?,” on pages 715–717.
This article contains supplemental material online at http://cjasn.asnjournals.org/lookup/suppl/doi:10.2215/CJN.10701016/-/DCSupplemental.
References
- 1.Shoskes DA, Halloran PF: Delayed graft function in renal transplantation: Etiology, management and long-term significance. J Urol 155: 1831–1840, 1996 [DOI] [PubMed] [Google Scholar]
- 2.Yarlagadda SG, Coca SG, Formica RN Jr, Poggio ED, Parikh CR: Association between delayed graft function and allograft and patient survival: A systematic review and meta-analysis. Nephrol Dial Transplant 24: 1039–1047, 2009 [DOI] [PubMed] [Google Scholar]
- 3.Lim WH, McDonald SP, Kennedy SE, Larkins N, Wong G: Association between slow and delayed graft function with graft outcomes in paediatric and adolescent deceased donor kidney transplant recipients [published online ahead of print August 29, 2016]. Transplantation doi: 10.1097/TP.0000000000001464 [DOI] [PubMed] [Google Scholar]
- 4.Bae S, Massie AB, Luo X, Anjum S, Desai NM, Segev DL: Changes in discard rate after the introduction of the kidney donor profile index (KDPI). Am J Transplant 16: 2202–2207, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Irish WD, Ilsley JN, Schnitzler MA, Feng S, Brennan DC: A risk prediction model for delayed graft function in the current era of deceased donor renal transplantation. Am J Transplant 10: 2279–2286, 2010 [DOI] [PubMed] [Google Scholar]
- 6.Balaz P, Rokosny S, Wohlfahrtova M, Wohlfahrt P, Bartonova A, Pokorna E, Honsova E, Viklicky O: Identification of expanded-criteria donor kidney grafts at lower risk of delayed graft function. Transplantation 96: 633–638, 2013 [DOI] [PubMed] [Google Scholar]
- 7.Bodonyi-Kovacs G, Putheti P, Marino M, Avihingsanon Y, Uknis ME, Monaco AP, Strom TB, Pavlakis M: Gene expression profiling of the donor kidney at the time of transplantation predicts clinical outcomes 2 years after transplantation. Hum Immunol 71: 451–455, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kotsch K, Kunert K, Merk V, Reutzel-Selke A, Pascher A, Fritzsche F, Tullius SG, Pratschke J: Novel markers in zero-hour kidney biopsies indicate graft quality and clinical outcome. Transplantation 90: 958–965, 2010 [DOI] [PubMed] [Google Scholar]
- 9.Wang CJ, Wetmore JB, Crary GS, Kasiske BL: The donor kidney biopsy and its implications in predicting graft outcomes: A systematic review. Am J Transplant 15: 1903–1914, 2015 [DOI] [PubMed] [Google Scholar]
- 10.Kainz A, Perco P, Mayer B, Soleiman A, Steininger R, Mayer G, Mitterbauer C, Schwarz C, Meyer TW, Oberbauer R: Gene-expression profiles and age of donor kidney biopsies obtained before transplantation distinguish medium term graft function. Transplantation 83: 1048–1054, 2007 [DOI] [PubMed] [Google Scholar]
- 11.Korbély R, Wilflingseder J, Perco P, Kainz A, Langer RM, Mayer B, Oberbauer R: Molecular biomarker candidates of acute kidney injury in zero-hour renal transplant needle biopsies. Transpl Int 24: 143–149, 2011 [DOI] [PubMed] [Google Scholar]
- 12.Anglicheau D, Suthanthiran M: Noninvasive prediction of organ graft rejection and outcome using gene expression patterns. Transplantation 86: 192–199, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartono C, Muthukumar T, Suthanthiran M: Noninvasive diagnosis of acute rejection of renal allografts. Curr Opin Organ Transplant 15: 35–41, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mannon RB: Immune monitoring and biomarkers to predict chronic allograft dysfunction. Kidney Int [Suppl 119]: S59–S65, 2010 [DOI] [PubMed] [Google Scholar]
- 15.Field M, Dronavalli V, Mistry P, Drayson M, Ready A, Cobbold M, Inston N: Urinary biomarkers of acute kidney injury in deceased organ donors--kidney injury molecule-1 as an adjunct to predicting outcome. Clin Transplant 28: 808–815, 2014 [DOI] [PubMed] [Google Scholar]
- 16.Cantaluppi V, Dellepiane S, Tamagnone M, Medica D, Figliolini F, Messina M, Manzione AM, Gai M, Tognarelli G, Ranghino A, Dolla C, Ferrario S, Tetta C, Segoloni GP, Camussi G, Biancone L: Neutrophil gelatinase associated lipocalin is an early and accurate biomarker of graft function and tissue regeneration in kidney transplantation from extended criteria donors. PLoS One 10: e0129279, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van den Akker EK, Hesselink DA, Manintveld OC, IJzermans JN, de Bruijn RW, Dor FJ: Neutrophil gelatinase-associated lipocalin, but not kidney injury marker 1, correlates with duration of delayed graft function. Eur Surg Res 55: 319–327, 2015 [DOI] [PubMed] [Google Scholar]
- 18.Dowling JK, O’Neill LA: Biochemical regulation of the inflammasome. Crit Rev Biochem Mol Biol 47: 424–443, 2012 [DOI] [PubMed] [Google Scholar]
- 19.Adib-Conquy M, Cavaillon JM: Stress molecules in sepsis and systemic inflammatory response syndrome. FEBS Lett 581: 3723–3733, 2007 [DOI] [PubMed] [Google Scholar]
- 20.Maxwell P: HIF-1: An oxygen response system with special relevance to the kidney. J Am Soc Nephrol 14: 2712–2722, 2003 [DOI] [PubMed] [Google Scholar]
- 21.Hsieh FY, Bloch DA, Larsen MD: A simple method of sample size calculation for linear and logistic regression. Stat Med 17: 1623–1634, 1998 [DOI] [PubMed] [Google Scholar]
- 22.de Jager W, Prakken BJ, Bijlsma JW, Kuis W, Rijkers GT: Improved multiplex immunoassay performance in human plasma and synovial fluid following removal of interfering heterophilic antibodies. J Immunol Methods 300: 124–135, 2005 [DOI] [PubMed] [Google Scholar]
- 23.Todd DJ, Knowlton N, Amato M, Frank MB, Schur PH, Izmailova ES, Roubenoff R, Shadick NA, Weinblatt ME, Centola M, Lee DM: Erroneous augmentation of multiplex assay measurements in patients with rheumatoid arthritis due to heterophilic binding by serum rheumatoid factor. Arthritis Rheum 63: 894–903, 2011 [DOI] [PubMed] [Google Scholar]
- 24.van den Ham HJ, de Jager W, Bijlsma JW, Prakken BJ, de Boer RJ: Differential cytokine profiles in juvenile idiopathic arthritis subtypes revealed by cluster analysis. Rheumatology (Oxford) 48: 899–905, 2009 [DOI] [PubMed] [Google Scholar]
- 25.Peters-Sengers H, Homan van der Heide JJ, Heemskerk MB, Ten Berge IJ, Ultee FC, Idu MM, Betjes MG, van Zuilen AD, Christiaans MH, Hilbrands LH, de Vries AP, Nurmohamed AS, Berger SP, Bemelman FJ: Similar 5-year estimated glomerular filtration rate between kidney transplants from uncontrolled and controlled donors after circulatory death-A dutch cohort study [published online ahead of print June 2, 2016]. Transplantation doi: 10.1097/TP.0000000000001211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rao PS, Schaubel DE, Guidinger MK, Andreoni KA, Wolfe RA, Merion RM, Port FK, Sung RS: A comprehensive risk quantification score for deceased donor kidneys: The kidney donor risk index. Transplantation 88: 231–236, 2009 [DOI] [PubMed] [Google Scholar]
- 27.Hall IE, Yarlagadda SG, Coca SG, Wang Z, Doshi M, Devarajan P, Han WK, Marcus RJ, Parikh CR: IL-18 and urinary NGAL predict dialysis and graft recovery after kidney transplantation. J Am Soc Nephrol 21: 189–197, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui LY, Zhu X, Yang S, Zhou JS, Zhang HX, Liu L, Zhang J: Prognostic value of levels of urine neutrophil gelatinase-associated lipocalin and interleukin-18 in patients with delayed graft function after kidney transplantation. Transplant Proc 47: 2846–2851, 2015 [DOI] [PubMed] [Google Scholar]
- 29.Parikh CR, Hall IE, Bhangoo RS, Ficek J, Abt PL, Thiessen-Philbrook H, Lin H, Bimali M, Murray PT, Rao V, Schroppel B, Doshi MD, Weng FL, Reese PP: Associations of perfusate biomarkers and pump parameters with delayed graft function and deceased-donor kidney allograft function. Am J Transplant 16(5): 1526–1539, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schröppel B, Krüger B, Walsh L, Yeung M, Harris S, Garrison K, Himmelfarb J, Lerner SM, Bromberg JS, Zhang PL, Bonventre JV, Wang Z, Farris AB, Colvin RB, Murphy BT, Vella JP: Tubular expression of KIM-1 does not predict delayed function after transplantation. J Am Soc Nephrol 21: 536–542, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Decruyenaere A, Decruyenaere P, Peeters P, Vermassen F: Validation in a single-center cohort of existing predictive models for delayed graft function after kidney transplantation. Ann Transplant 20: 544–552, 2015 [DOI] [PubMed] [Google Scholar]
- 32.Gourishankar S, Grebe SO, Mueller TF: Prediction of kidney graft failure using clinical scoring tools. Clin Transplant 27: 517–522, 2013 [DOI] [PubMed] [Google Scholar]
- 33.Le Dinh H, Weekers L, Bonvoisin C, Krzesinski JM, Monard J, de Roover A, Squifflet JP, Meurisse M, Detry O: Delayed graft function does not harm the future of donation-after-cardiac death in kidney transplantation. Transplant Proc 44: 2795–2802, 2012 [DOI] [PubMed] [Google Scholar]
- 34.Sánchez-Fructuoso A, Prats Sánchez D, Marqués Vidas M, López De Novales E, Barrientos Guzmán A: Non-heart beating donors. Nephrol Dial Transplant 19[Suppl 3]: iii26–iii31, 2004 [DOI] [PubMed] [Google Scholar]
- 35.Heilman RL, Smith ML, Smith BH, Qaqish I, Khamash H, Singer AL, Kaplan B, Reddy KS: Progression of interstitial fibrosis during the first year after deceased donor kidney transplantation among patients with and without delayed graft function. Clin J Am Soc Nephrol 11: 2225–2232, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bestard O, Cruzado JM, la Franquesa M, Grinyó JM: Biomarkers in renal transplantation. Curr Opin Organ Transplant 15: 467–473, 2010 [DOI] [PubMed] [Google Scholar]
- 37.Zaza G, Ferraro PM, Tessari G, Sandrini S, Scolari MP, Capelli I, Minetti E, Gesualdo L, Girolomoni G, Gambaro G, Lupo A, Boschiero L: Predictive model for delayed graft function based on easily available pre-renal transplant variables. Intern Emerg Med 10: 135–141, 2014 [DOI] [PubMed] [Google Scholar]
- 38.Lin YH, Huang CJ, Chao JR, Chen ST, Lee SF, Yen JJ, Yang-Yen HF: Coupling of osteopontin and its cell surface receptor CD44 to the cell survival response elicited by interleukin-3 or granulocyte-macrophage colony-stimulating factor. Mol Cell Biol 20: 2734–2742, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao J, Chen L, Shu B, Tang J, Zhang L, Xie J, Qi S, Xu Y: Granulocyte/macrophage colony-stimulating factor influences angiogenesis by regulating the coordinated expression of VEGF and the Ang/Tie system. PLoS One 9: e92691, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fernández-Riejos P, Najib S, Santos-Alvarez J, Martín-Romero C, Pérez-Pérez A, González-Yanes C, Sánchez-Margalet V: Role of leptin in the activation of immune cells. Mediators Inflamm 2010: 568343, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Procaccini C, Jirillo E, Matarese G: Leptin as an immunomodulator. Mol Aspects Med 33: 35–45, 2012 [DOI] [PubMed] [Google Scholar]
- 42.Mancuso P, Myers MG Jr, Goel D, Serezani CH, O’Brien E, Goldberg J, Aronoff DM, Peters-Golden M: Ablation of leptin receptor-mediated ERK activation impairs host defense against gram-negative pneumonia. J Immunol 189: 867–875, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Y, Yu X, Chen H, Sjöberg S, Roux J, Zhang L, Ivoulsou AH, Bensaid F, Liu CL, Liu J, Tordjman J, Clement K, Lee CH, Hotamisligil GS, Libby P, Shi GP: Leptin deficiency shifts mast cells toward anti-inflammatory actions and protects mice from obesity and diabetes by polarizing M2 macrophages. Cell Metab 22: 1045–1058, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Jager W, te Velthuis H, Prakken BJ, Kuis W, Rijkers GT: Simultaneous detection of 15 human cytokines in a single sample of stimulated peripheral blood mononuclear cells. Clin Diagn Lab Immunol 10: 133–139, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.