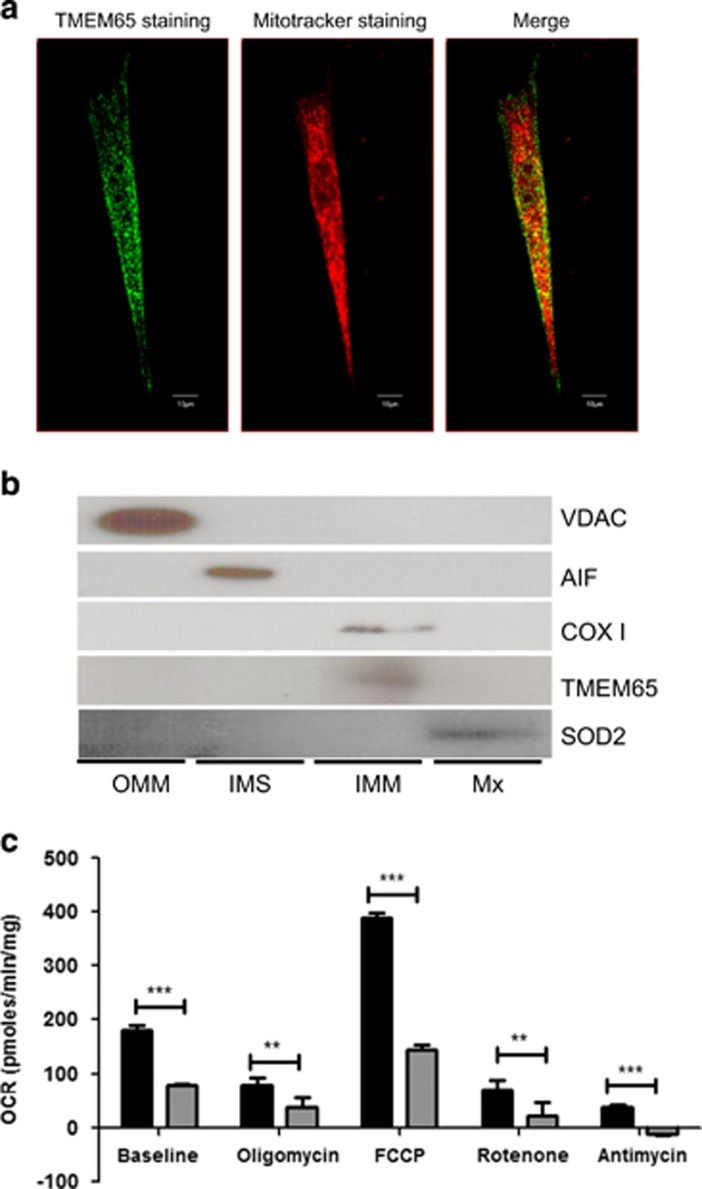
Figure 2.
TMEM65 localizes to the inner mitochondrial membrane and a mutation in TMEM65 effects cellular respiration. (a) Dermal fibroblasts taken from healthy control were stained for TMEM65 protein (green) and mitochondria (red) and images were taken on an inverted confocal microscope (Olympus). Co-localization of TMEM65 protein (green) and Mitotracker deep red was captured and calculated by Olympus Fluoview software Version 4.2 (scale bar=10 μm). The yellow color is reflective of the co-localization. (b) Mitochondria isolated from skeletal muscle were subfractionated into the outer mitochondrial membrane (OMM), intermembrane space (IMS), inner mitochondrial membrane (IMM) and matrix (Mx) fractions, and these fractions were subsequently immunoblotted for the compartment-specific proteins voltage-dependent anion channel (VDAC), apoptosis-inducing factor (AIF), cytochrome c oxidase subunit I (COX I), transmembrane protein 65 (TMEM65) and superoxide dismutase 2 (SOD2). (c) Oxygen consumption rate in primary dermal fibroblasts from a TMEM65 patient (gray bars) and compared with primary dermal fibroblast from three healthy age-matched controls (black bars). Oxygen consumption rate (OCR) was determined using the XFe24 Extracellular Flux Analyzer (Seahorse Bioscience, Billerica, MA, USA). During oxygen consumption rate measurements cells were exposed to metabolic inhibitors including oligomycin, FCCP, rotenone and antimycin. The dermal fibroblast from the TMEM65 patient showed an overall reduced cellular respiration rates. The values are normalized to the total cellular protein per well. Graph represents three individual experiments. **P<0.001, ***P<0.0001.