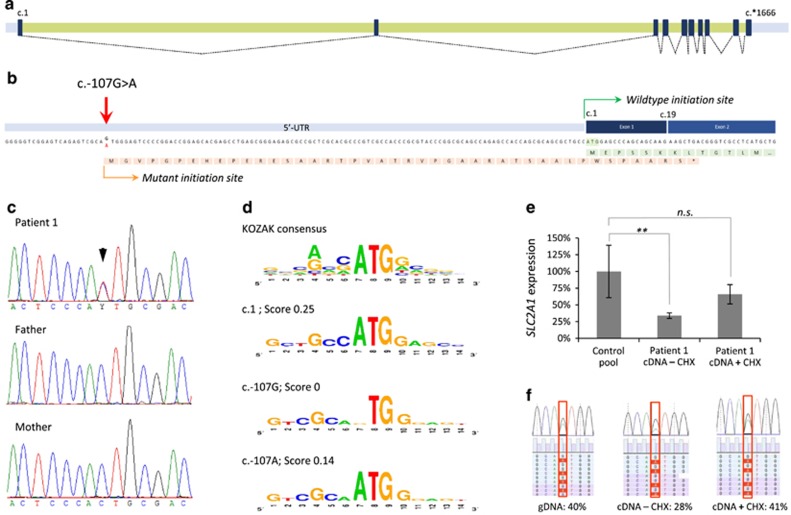
Figure 1.
Molecular characterization and expression analysis of the SLC2A1 c.-107G>A variant. (a) Schematic depiction of the SLC2A1 exons (black boxes), introns (green bars), and UTRs (gray bars) with splicing information (dotted lines) based on NM_006516.2. (b) The c.-107G>A variant results in a potential novel translation initiation site. The predicted protein effect is indicated in orange, whereas the wild-type protein (partial) is shown in green. The mutant protein is predicted to result in a premature termination codon in exon 2 (*). (c) Sanger sequencing validation of the c.-107G>A variant in patient 1 and her parents, indicating de novo occurrence of the variant. (d) Evaluation of the sequence context surrounding c.107G for the presence of a KOZAK sequence. Top panel shows the consensus sequence, with the relative height of the nucleotides representing the frequency observed at the specified position. Second panel shows the wild-type SLC2A1 c.1 KOZAK sequence. Third panel shows the predication for the wild-type c.107G sequence and its absence of a KOZAK sequence. Lower panel shows the prediction for c.107 A with the presence of a KOZAK sequence. Scores represent reliability of the KOZAK prediction. (e) Relative SLC2A1 mRNA expression in patient 1, indicating a significantly reduced expression (cDNA – CHX) that is partially increased by blocking of nonsense-mediated decay with cycloheximide (cDNA+CHX); **P<0.01. (f) Nonsense-mediated decay of the SLC2A1 transcript bearing the c.-107G>A variant. Patient's fibroblasts were cultured in the absence (−CHX) or presence (+CHX) of cycloheximide, and both genomic (gDNA) and mRNA were isolated. Copy-DNA (cDNA) was made from the mRNA samples, and both gDNA and cDNA were subjected to semi-quantitative ion-torrent sequencing (>40 000 reads per sample) after PCR. The percentage of reads containing the c.-107G>A variant is given. NS, not significant.