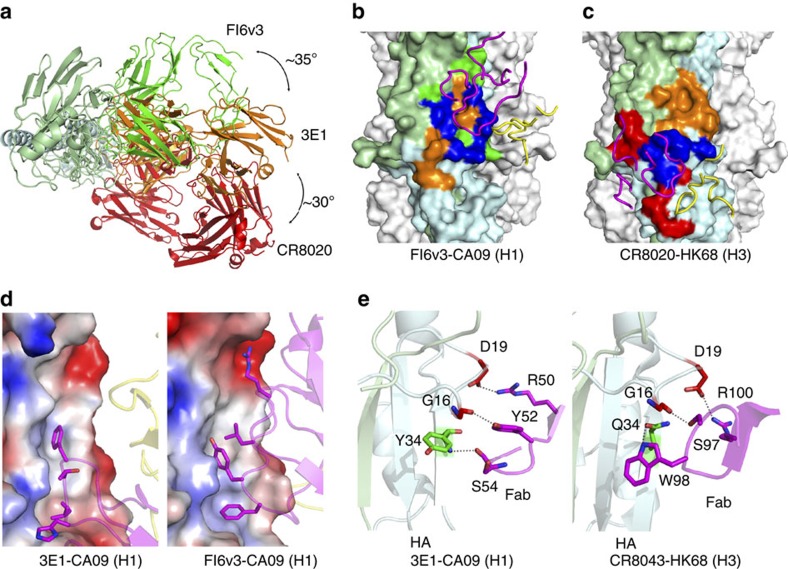
Figure 4. Binding specificities of 3E1 compared with the type I and type II bnmAbs.
(a) Top view overlay of the crystal structures of the 3E1 (orange), FI6v3 (green, type I bnmAb) and CR8020 (red, type II bnmAb) Fabs in complexes with HA. For clarity, only the HA in the 3E1 Fab–CA09 HA complex is shown. The three Fabs bind to HA from different orientations. (b) Comparison of the epitopes between 3E1 and FI6v3. (c) Comparison of the epitopes between 3E1 and CR8020. The HAs in the FI6v3 Fab-CA09 HA complex (PDB code: 3ZTN) and CR8020 Fab-HK68 HA complex (PDB code: 3SDY) are shown in surface representation and the CDRs of FI6v3 and CR8020 in cartoons. The epitopes of 3E1, FI6v3 and CR8020 are coloured in orange, green and red, respectively, and the overlapping regions are coloured in blue. (d) Similar to FI6v3 (type I bnmAbs), 3E1 Fab makes hydrophobic contacts with the F subdomain of HA. The HAs are shown as electrostatic surface representations and the Fabs as cartoons in the same orientation. (e) Similar to CR8043 (type II bnmAb), 3E1 makes hydrophilic interactions with the fusion peptide of HA. The residues of HAs and Fabs involved in the interactions are shown with side chains.