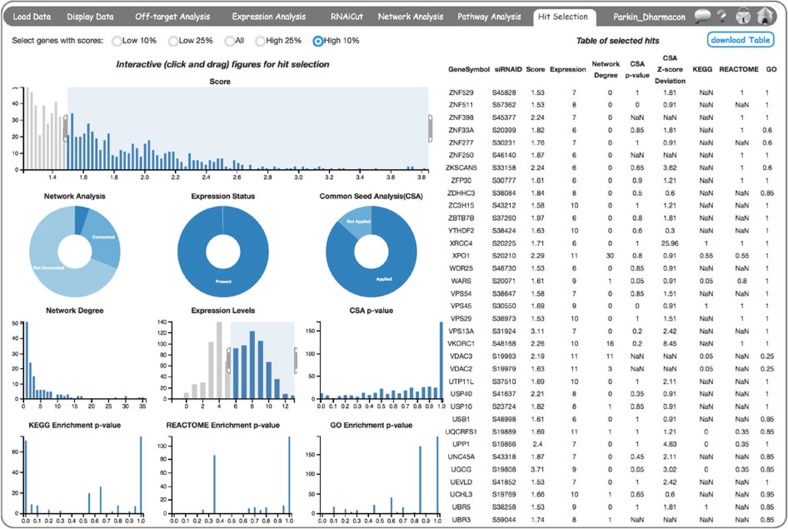
Figure 8. CARD Hit Selection page.
Hit Selection page providing both a summary and details of the hit selection process. Choosing a fraction of the genes (10%) from the top radio buttons expedites data loading from genome-scale screens. On the left side, a summary of the results is shown as interactive bar and pie charts. On the top row, a bar chart shows the histogram of RNAi screen scores. The Network Analysis pie chart shows fraction of genes connected, not connected and unknown as obtained from the Network Analysis page, whereas the Network Degree histogram shows the degree distribution of the connected genes. Similarly, the gene Expression Status pie chart shows the fraction of present/absent genes and the Expression Levels histogram shows an often-bimodal distribution of expression, where the left mode corresponds to likely absent genes. The common seed analysis (CSA) pie chart shows the fraction of siRNAs for which CSA was applied and not applied, then the CSA P-value distribution is shown specifically for the applied subset. As KEGG, Reactome and GO databases have information for only a subset of the genome, the histograms show the enrichment P-values of the selected genes present in those databases. Mouse-over on chart headers provides description of the chart. On the right panel, an output table of the selected hits is provided. Each of the charts is interactive, such that clicking and dragging to select a subset of genes applies filters that are dynamically reflected in the table of hit genes.