Figure 1.
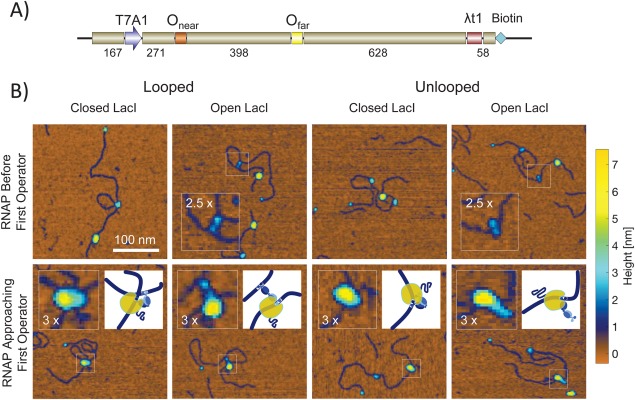
Nanographs of RNA polymerases trapped by EDTA quenching during elongation along DNA with and without LacI‐mediated loops. (A) Schematic representation of the DNA templates used in scanning force microscopy assays. All templates contained a T7A1 promoter close to the upstream end, a stall site at position +22, a “far” O1 operator, the lambda t1 terminator (λt1) and a biotin label at the downstream end. The two DNA templates used for SFM measurements of transcription differed in the “near” operator positioned 253 bp downstream from the promoter; one template contained the Os operator while the other contained the O2 operator. The terminator was the very last feature of the sequence and was biotin labeled. Streptavidin was coupled to the biotin label to facilitate identifying the “downstream” end of the molecule in SFM nanographs. (B) The upper row is a selection of molecules along which RNA polymerases (large yellow particle) had not progressed very far from the transcription start site near the end of the DNA without a streptavidin particle (blue). Closed and open conformations of the LacI tetramers are visible for either looped (left) or unlooped (right) columns. The closed conformations are shown as blue particles that are slightly larger than the streptavidin. In the open conformation, two lobes are visible especially on looped DNA. These lobes correspond to individual dimers with DNA binding head groups. The TECs shown in the lower row had progressed further and small coils of RNA emanate from them (see inset schematics for the regions of interest). These TECs have collided with LacI particles. The LacI particles correspond to blue protuberances on the periphery of the larger yellow RNA polymerase particle. The RNA polymerases themselves appear to shift to the side opposite LacI especially for open LacI conformations.
