Figure 3.
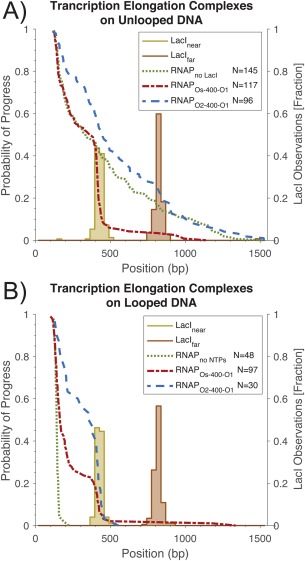
The probability of progress through obstacles. The positions of RNA polymerases and lac repressor molecules were measured along DNA contours in scanning force microscopy images. The probability of progress for each position was calculated as 1‐ [normalized cumulative histogram of RNAP position]. (See Supporting Information Fig. S2 for standard histograms of RNAP positions.) (A) Unlooped DNA templates. Without LacI obstacles (green dotted line), the majority of RNA polymerases began to transcribe (transcription elongation complexes, TECs) and after 60 s were distributed randomly along unlooped DNA templates. When LacI obstacles were present, a high percentage of TECs were blocked by LacI bound to the high affinity, Os operator (red, short‐dashed line). In contrast, a large percentage of TECs surpassed LacI bound to the lower affinity, O2 operator (blue, long‐dashed lines). (B) Looped DNA templates. Without NTPs, RNA polymerases were tightly bunched at the promoter site on DNA molecules with LacI‐mediated loops (green dotted line). Almost all TECs were blocked by LacI mediating loops between O1 and the high affinity, Os operator (red, short‐dashed line). Remarkably, LacI tetramers securing loops between O1 and the lower affinity, O2 operator, were equally effective obstacles (blue long‐dashed line). In each graph the yellow and orange histograms indicate the distributions measured for LacI along the DNA contours.
