Figure 16.
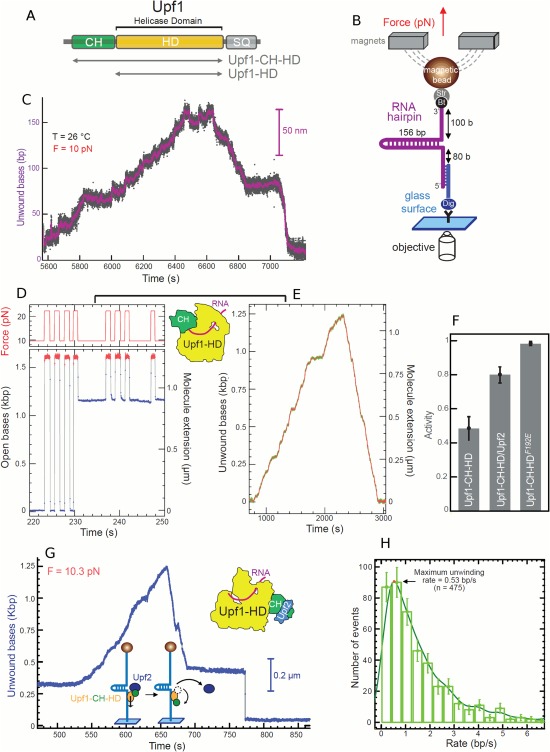
Upf1 helicase activity on RNA. (A) Organization of Upf1 and truncated versions (gray arrows) used in our study, Upf1‐HD and Upf1‐CH‐HD. (B) Schematic representation of the RNA used for the MT set up. (C) Experimental trace showing the activity of Upf1‐HD in saturating concentration of ATP. The number of unwound bases is deduced from the molecular extension Z(t) obtained at F = 10pN. From 5600 s to 6440 s, the helicase unwound the 156bp RNA hairpin. From 6640 s to 7200 s, the RNA hairpin refolded, while Upf1‐HD translocated on ssRNA reaching the 3′extremity. (D, E, and F) The binding of Upf2 to CH domain activates Upf1‐CH‐HD unwinding and translocation. (D, E) Experimental traces corresponding to the two types of enzymatic activity detected for Upf1 on a DNA hairpin. (D) Upf1 binds on ssDNA (starting at 231 s), blocking the re‐zipping of the hairpin. (E) The enzyme is active. (F) Histogram of relative activity of Upf1‐CH‐HD, Upf1‐CH‐HD/Upf2 complex and Upf1F192E mutant. (G) Trace of human Upf1‐CH‐HD/Upf2 complex unwinding steadily and completely the DNA molecule, passing the apex, pursuing its translocation and refolding the DNA hairpin. At t = 675 s, the complex makes two strand switching events before finally stopping its activity leaving the hairpin blocked (at t = 687 s). This trace is atypical since the Upf1‐CH‐HD/Upf2 complex translocates for some time before stalling; for most bursts, the complex stops very quickly after passing the apex. (H) Distribution of instantaneous unwinding rate of the Upf1‐CH‐HD/Upf2 complex.87
