Figure 6.
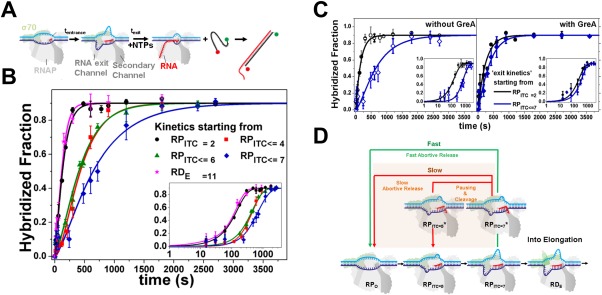
Paused backtracked intermediate in RNAP initiation. Pausing in initiation by RPITC is stabilized by backtracking. Reprinted from Lerner and Chung et al.90 with permission. (A) Schematic of transcription kinetics starting from particular NTP starved ITC states (incubation with a partial set of NTPs for tentrance) using single‐round quenched kinetics assays. Upon adding all NTPs, transcription kinetics start and stop at different incubation times (texit). Transcripts were quantified via hybridization to a ssDNA FRET probe. (B) Quenched kinetics results identify an initiation‐related stalled state. Shown are run‐off kinetics from various NTP‐starved states. Kinetics starting from late initiation states (e.g., RPITC ≤ 7, blue) are slower than from an earlier initiation state (e.g., RPITC = 2, black). (C) GreA suppresses the kinetic delay in transcription initiation. Left; Run‐off transcription kinetics are slower when starting from RPITC ≤ 7 (blue) than from RPITC = 2 (black). Right; with 1 µM GreA, the delay in transcription initiation is reduced. All data are represented as points and solid lines represent best‐fit as described in90. (D) A modified transcription initiation model. RNAP transcription initiation branches to promoter clearance and transitions into elongation (black arrows) or into release of abortive transcripts (green and red arrows). After initial backtracking steps (e.g., from RPITC = 7 to RPITC = 7*), the complex can continue with either fast abortive transcript release (classic model, green arrow) or transition into a paused‐backtracked state. Exit from the paused–backtracked state can occur either by successive slow backtracking steps (red arrow) or through intrinsic cleavage of RNA bases in the secondary channel, which prepares RNAP in, e.g., the RPITC = 5 state. Upon cleavage, the complex can release the abortive transcript or re‐establish RNA polymerization (e.g., from the RPITC = 5 state).
