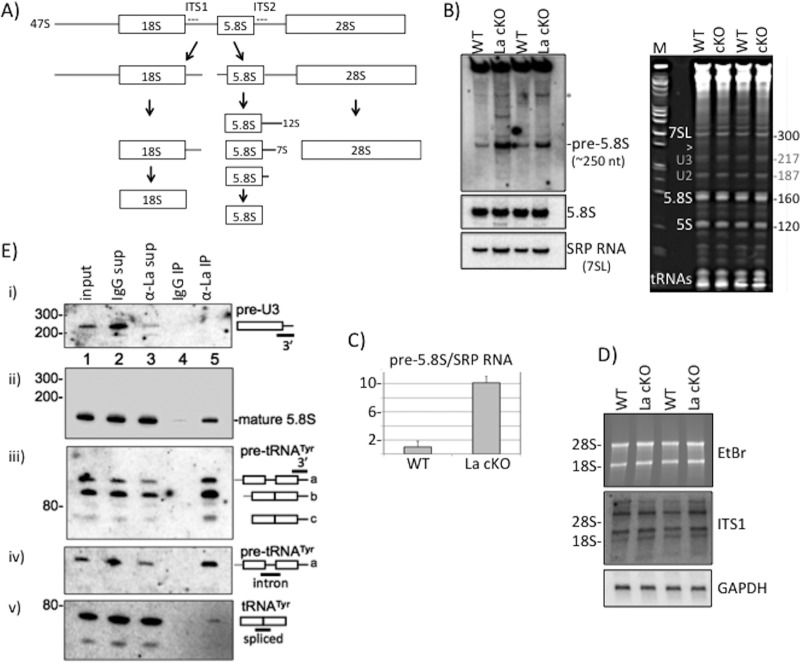
FIG 1.
Pre-rRNA processing perturbation in La cKO cortex. (A) Simplified diagram of the rRNA processing pathway in mice (see the text). The locations of the ITS1 and ITS2 probes used for Northern blot analyses are indicated with dashed lines. (B) (Left) Northern blot of 13-week cortex total RNA using an ITS2 probe revealing the pre-5.8S rRNA species (see the text) (top), a probe to mature 5.8S rRNA (middle), and a probe to SRP 7SL RNA (bottom). (Right) Ethidium bromide (EtBr)-stained gel used to produce the blot. The arrowhead indicates the approximate position of the pre-5.8S rRNA relative to the 7SL RNA of 300 nt; the bands annotated with gray numbers are believed to be U2 and U3 snoRNAs. (C) Quantification of the pre-5.8S rRNA band relative to the SRP RNA bands using the following formula: La cKO(pre-5.8S/SRP)/WT(pre-5.8S/SRP), with WT set to 1. The error bars reflect two biological replicates. (D) Northern blot hybridization after agarose gel electrophoresis (EtBr-stained gel [top]) using an ITS1 probe (middle) and a GAPDH probe used as a loading control (bottom). The positions of the 28S and 18S rRNAs are indicated. (E) Analysis of La-associated RNAs by IP of mouse cortex lysate with anti-mLa or control IgG. Ten percent of inputs and supernatants were electrophoresed in a 10% TBE-urea gel, along with 100% of the bead-bound eluate RNA from the IPs, as indicated above the lanes. Each of the gels (i to v) shows the results of the same blot after hybridization with a different probe, as indicated on the right and in the text.