FIG 3.
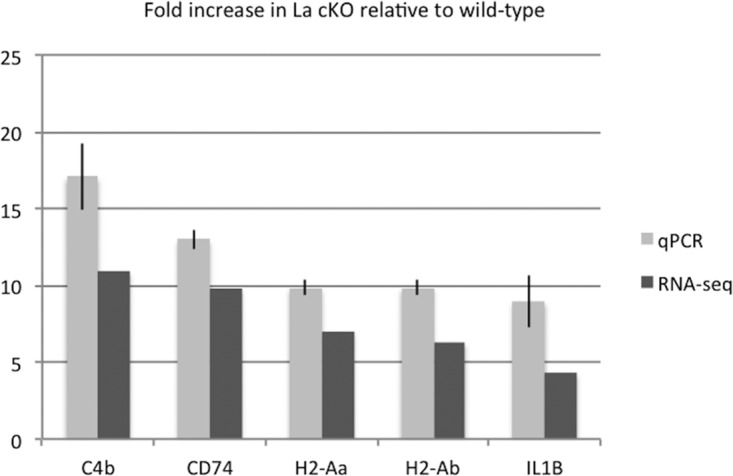
Agreement between qPCR and RNA-Seq results. Table 1 lists the mRNAs found by RNA-Seq to be significantly altered in abundance in La cKO relative to WT cortex. To validate and determine the accuracy of the RNA-Seq data, 5 mRNAs were chosen that were either directly involved in immune cell activation (C4b, H2-Aa, and H2-Ab) or implicated in neurodegenerative pathologies (CD74 and IL-1β). qPCR was performed on two La cKO and two WT biological replicate samples. Threshold cycle (CT) values were normalized to GAPDH, and the fold change was calculated relative to the WT samples; relative expression levels were calculated using the 2−ΔΔCT method (157). The error bars indicate 95% confidence intervals for both RNA-Seq and qRT-PCR data. For each mRNA, a negative-control reaction omitting reverse transcriptase showed negligible amplification (≤2%), indicating dependence on input RNA (not shown).
