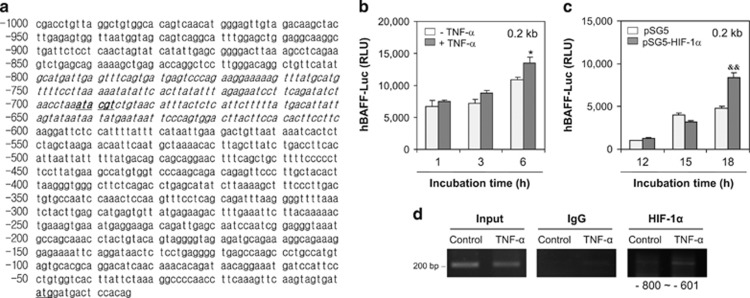
Figure 7.
A 0.2 kb hBAFF promoter (−800 to −601 bp) activity was enhanced by TNF-α treatment and HIF-1α binding. (a) Transcription of hBAFF gene started at the site of bold nucleotide with underline on promoter. Italic bold nucleotide with underline showed HIF-1α binding site on hBAFF promoter. The 0.2 kb (italic nucleotides, −800 to −601 bp) were cloned into pGL3 plasmids (0.2 kb pGL3-hBAFF-Luc). (b and c) MH7A cells were transfected with 0.2 kb pGL3-hBAFF-Luc plasmid. Then, the cells were incubated in the presence or absence of TNF-α for 6 h (b). MH7A cells were co-transfected with 0.2 kb pGL3-hBAFF-Luc and pSG5-HIF-1α plasmids (c). Luciferase activity of hBAFF promoter was measured by using luminometer. Data were the representative of four experiments. Data in the bar graph represent the means±S.E.M. *P<0.05, significant difference as compared with TNF-α-untreated control at each time point (b). &&P<0.01, significant difference as compared with pSG5 plasmid-transfected control at each time point (c). MH7A cells were stimulated with TNF-α and fixed with 1% formaldehyde. Their chromatin extracts were immunoprecipitated with anti-HIF-1α antibodies. DNA fragments were subjected to PCR analysis using primer sets spanning the promoter regions. Sequences for primer set were 5′-GCATGATTGAGTTTCAGTGA-3′ (forward) and 5′-GAAHGAAGTGTGGAAGTAAG-3′ (reverse). Primer set corresponds to −800 to −601 bp including HIF-1α binding site (−693 to −688 bp) on hBAFF promoter. Data were the representative of four experiments (d)