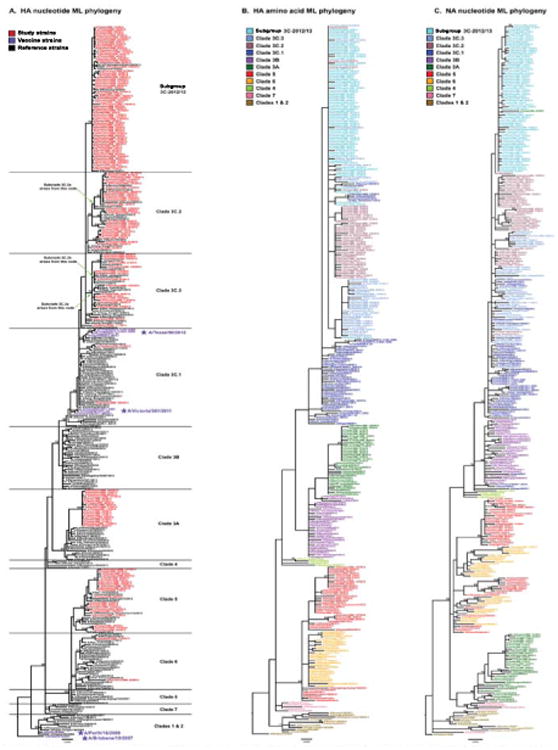
Figure 1. Phylogenetic relationships among the influenza A(H3N2) study samples, vaccine strains, and selected references inferred from maximum likelihood analyses of the HA nucleotide (A), HA protein (B), and NA nucleotide (C) sequences, Texas, 3 November 2012-8 February 2013 (n = 154).
CDC: Centers for Disease Control and Prevention; HA: haemagglutinin; ML: maximum likelihood; NA: neuraminidase; WHO: World Health Organization
Bootstrap values for nodes with ≥70% support following 500 replicates are provided. Clades are classified following the CDC nomenclature [25] as closely as possible and are defined based on the HA nucleotide phylogeny (A). A new subgroup, designated 3C-2012/13, appears to have arisen during the 2012/13 influenza season. The nodes from which the new WHO-designated 3C.2a, 3C.3a, and 3C.3b subclades subsequently arise are marked on the phylogeny to place our 2012/13 study in the context of more recent seasons; placement was based on additional ML and neighbour-joining phylogenetic analyses using available H3 sequence data through 2014 (data not shown). For the HA protein (B) and NA nucleotide phylogenies (C), strain names are coloured by HA nucleotide clade definitions, showing that the HA protein and NA nucleotide clades largely match the nucleotide-based HA clade definitions. However, some interleaving of 3C subclades is observed in the HA protein tree, indicating that these subclades diverge largely through synonymous nucleotide mutations. The interleaving of colours in the NA nucleotide tree signifies intrasubtypic reassortment between the HA and NA gene segments. Scale bars indicate the average number of nucleotide changes per site.
The authors gratefully acknowledge the 87 originating and submitting laboratories who directly contributed sequences used in the phylogenetic analyses to GISAID: ADImmune Corporation, Taiwan; Alabama Department of Public Health, Bureau of Clinical Laboratories; Arkansas Children's Hospital; Austin Health, Australia; California Department of Health Services; Canterbury Health Services, New Zealand; US Centers for Disease Control and Prevention; CRR virus Influenza region Sud, France; Delaware Public Health Lab; Gart Naval General Hospital, United Kingdom; Georgia Public Health Laboratory; Government Virus Unit, Hong Kong; Health Protection Agency, England; Health Protection Inspectorate, Estonia; Hospital Clinic, Spain; Institute of Epidemiology and Infectious Diseases AMS of Ukraine; Institute of Epidemiology Disease Control and Research (IEDCR) & Bangladesh National Influenza Centre (NIC); Institute of Immunology and Virology Torlak, Serbia; Institute of Medical and Veterinary Science (IMVS), Australia; Institute of Public Health, Montenegro; Institute de Salud Publica de Chile; Institute Nacional de Saude, Portugal; Institut Pasteur de Dakar, Senegal; Institut Pasteur de Madagascar; Iowa State Hygienic Laboratory; Istituto Superiore di Sanit, Italy; Kansas Department of Health and Environment; Kentucky Division of Laboratory Services; Laboratory for Virology, National Institute of Public Health, Slovenia; Laboratory of Influenza and ILI, Belarus; Landspitali - University Hospital, Iceland; Louisiana Department of Health and Hospitals; Maine Health and Environmental Testing Laboratory; Maryland Department of Health and Mental Hygiene; Melbourne Pathology, Australia; Michigan Department of Community Health; Ministry of Health of Ukraine; Minnesota Department of Health; Monash Medical Centre, Australia; Montana Laboratory Services Bureau; Montana Public Health Laboratory; National Centre of Infectious and Parasitic Diseases, Bulgaria; National Institute for Communicable Disease, South Africa; National Institute for Health and Welfare, Finland; National Institute for Medical Research, United Kingdom; National Institute of Health, Korea; National Microbiology Laboratory, Health Canada; Nebraska Public Health Lab; Nevada State Health Laboratory; New Hampshire Public Health Laboratories; New Jersey Department of Health & Senior Services; New York City Department of Health; New York Medical College; New York State Department of Health; Norwegian Institute of Public Health; Oregon Public Health Laboratory; Pathwest QE II Medical Centre, Australia; Pennsylvania Department of Health; Prince of Wales Hospital, Australia; Queensland Health Scientific Services, Australia; Republic Institute for Health Protection, Macedonia; Rhode Island Department of Health; Robert Koch-Institute, Germany; San Antonio Metropolitan Health, Texas; South Dakota Public Health Lab; Southern Nevada Public Health Lab; Spokane Regional Health District, Washington; Statens Serum Institute, Denmark; State of Hawaii Department of Health; State of Idaho Bureau of Laboratories; Swedish Institute for Infectious Disease Control; Texas Department of State Health Services-Laboratory Services; University of Vienna, Austria; U.S. Air Force School of Aerospace Medicine; Utah Department of Health; VACSERA, Egypt; Victorian Infectious Diseases Reference Laboratory, Australia; Virginia Division of Consolidated Laboratories; Westmead Hospital, Australia; West Virginia Office of Laboratory Services; WHO Chinese National Influenza Center; WHO Collaborating Centre for Reference and Research on Influenza, Australia; WHO National Influenza Centre, Norway; WHO National Influenza Centre, National Institute of Medical Research (NIMR), United Kingdom; WHO National Influenza Centre Russian Federation; Wisconsin State Laboratory of Hygiene; and Wyoming Public Health Laboratory.
