Figure 3. Genome constellations for coding-complete influenza A(H3N2) sequences from the 2012/13 influenza epidemic for all clades (A) and for clade 3 (B), Texas, 3 November 2012–8 February 2013 (n =154).
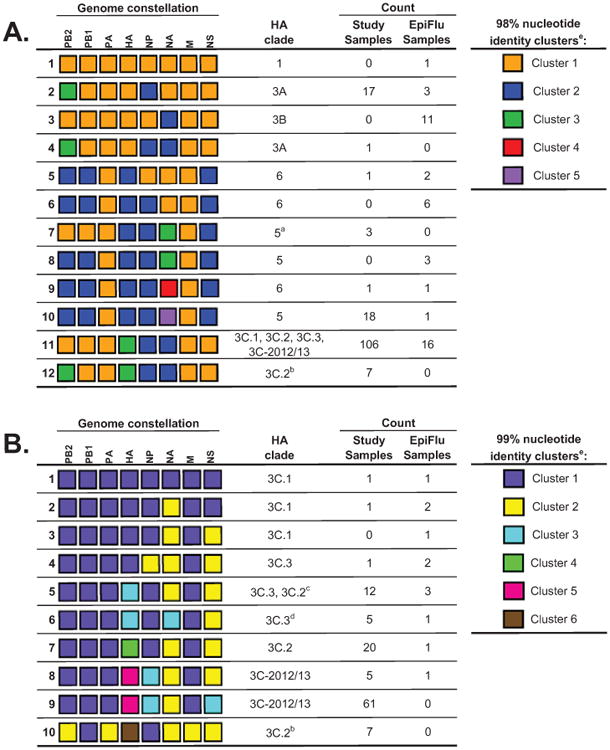
a These study samples form their own HA monophyly within clade 5.
b These study samples form their own HA monophyly within clade 3C.2.
c Of the 13 strains in this constellation that are also in the HA phylogeny, only one strain falls within clade 3C.2.
d These study samples form their own HA monophyly within clade 3C.3.
e Please note that each column is evaluated independently and cluster numbering is arbitrary.
- Genome constellation 1 (orange) segments were defined as sharing 98% nucleotide identity with the A/Perth/16/2009 vaccine strain. Twelve unique genome constellations were identified at the 98% nucleotide identity cut-off, eight of which were observed in the study samples.
- Clade 3 genome constellation 1 (purple) segments were defined as sharing 99% nucleotide identity with the A/Victoria/361/2011 vaccine strain. 10 unique genome constellations were identified at the 99% nucleotide identity cut-off, nine of which were observed in the study samples.
The authors gratefully acknowledge the 32 originating and submitting laboratories who directly contributed sequences used in the constellation analysis to GISAID: Alaska State Virology Lab; Arizona Department of Health Services; Austin Health, Australia; California Department of Health Services; Canterbury Health Services, New Zealand; US Centers for Disease Control and Prevention; Institute of Medical and Veterinary Science (IMVS), Australia; Institut Pasteur New Caledonia; Iowa State Hygienic Laboratory; John Hunter Hospital, Virology Unit, Clinical Microbiology, Australia; Kentucky Division of Laboratory Services; Melbourne Pathology, Australia; Michigan Department of Community Health; New Mexico Department of Health; New York State Department of Health; Papua New Guinea Institute of Medical Research; Pathwest QE II Medical Centre, Australia; Pennsylvania Department of Health; Puerto Rico Department of Health; Queensland Health Scientific Services, Australia; Research Institute of Tropical Medicine, Philippines; Rhode Island Department of Health; Royal Hobart Hospital, Australia; Southern Nevada Public Health Lab; Spokane Regional Health District, Washington; State of Hawaii Department of Health; Texas Department of State Health Services-Laboratory Services; USAMC-AFRIMS Department of Virology, Cambodia; Utah Department of Health; Victorian Infectious Diseases Reference Laboratory, Australia; WHO Collaborating Centre for Reference and Research on Influenza, Australia; and WHO National Influenza Centre, National Institute of Medical Research (NIMR), United Kingdom.
