Figure 4. Location of antigenic sites on the H3 monomer, along with key clade 3C substitutions and glycosylation sites.
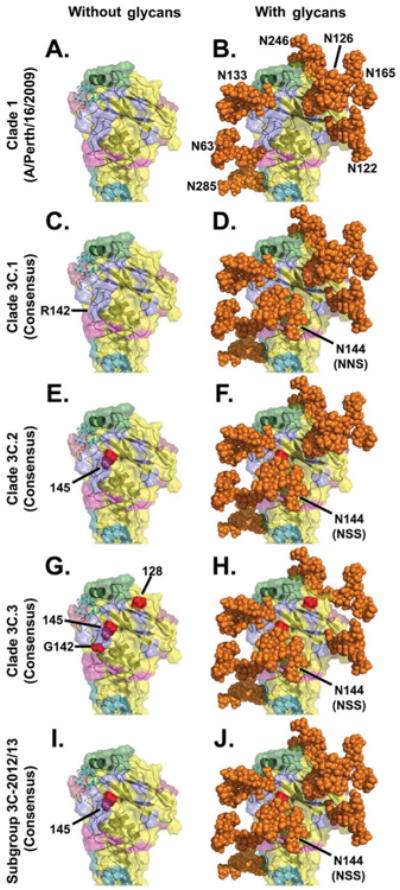
In all panels, the peptide backbone of the HA globular head is represented as a ribbon with a translucent solid surface using the A/Finland/486/2004 HA crystal structure, 2YP3, bound to a synthetic 2,6-sialic acid ligand (cyan stick structure) [40]. In panels on the right, all potential N-linked glycans in the H3 globular head were modelled using the AllosMod server [41] and rendered by PyMOL [45] as solid orange spheres.
Panels A and B: A representation of A/Perth/16/2009 (clade 1) illustrating previously defined H3 antigenic sites [60-63]: lavender (antigenic site A), green (antigenic site B), deep teal (antigenic site C), raspberry (antigenic site D), and light magenta (antigenic site E). A/Perth/16/2009 lacks the N144 glycosylation site that all subclade/subgroup 3C viruses have.
Panels C and D: A representation of 3C.1 viruses illustrating the H3 antigenic sites and the presence of the NNS sequon that may allow for glycosylation at N144, possibly at a reduced efficiency [55].
Panels E–J: Critical amino acid differences between the 3C.1 consensus and the indicated 3C subclade/subgroup consensuses are shown in red and labelled with H3 structural numbering.
Panel G: 3C.3 viruses have an R142G substitution, which removes most of the bulk of that protruding side chain; in this panel, the structure was modified to remove the bulky arginine (R) side chain (cf. the small red spheres for residue 142 with the larger lavender protrusion that extends downward in the other panels, as labelled in C).
Panel F: Some 3C.2 viruses have lost the N122 glycan.
Panel H: 3C.3 viruses lack the N126 glycan.
Panel J: 3C-2012/13 viruses are among the most heavily glycosylated.
