Abstract
Purpose
In squamous cell carcinomas of the head and neck (HNSCC), the increasing incidence of oropharyngeal squamous cell carcinomas (OPSCCs) is attributable to human papillomavirus (HPV) infection. Despite commonly presenting at late stage, HPV-driven OPSCCs are associated with improved prognosis compared with HPV-negative disease. HPV DNA is also detectable in nonoropharyngeal (non-OPSCC), but its pathogenic role and clinical significance are unclear. The objectives of this study were to determine whether HPV plays a causal role in non-OPSCC and to investigate whether HPV confers a survival benefit in these tumors.
Methods
Meta-analysis was used to build a cross-tissue gene-expression signature for HPV-driven cancer. Classifiers trained by machine-learning approaches were used to predict the HPV status of 520 HNSCCs profiled by The Cancer Genome Atlas project. DNA methylation data were similarly used to classify 464 HNSCCs and these analyses were integrated with genomic, histopathology, and survival data to permit a comprehensive comparison of HPV transcript-positive OPSCC and non-OPSCC.
Results
HPV-driven tumors accounted for 4.1% of non-OPSCCs. Regardless of anatomic site, HPV+ HNSCCs shared highly similar gene expression and DNA methylation profiles; nonkeratinizing, basaloid histopathological features; and lack of TP53 or CDKN2A alterations. Improved overall survival, however, was largely restricted to HPV-driven OPSCCs, which were associated with increased levels of tumor-infiltrating lymphocytes compared with HPV-driven non-OPSCCs.
Conclusion
Our analysis identified a causal role for HPV in transcript-positive non-OPSCCs throughout the head and neck. Notably, however, HPV-driven non-OPSCCs display a distinct immune microenvironment and clinical behavior compared with HPV-driven OPSCCs.
INTRODUCTION
High-risk human papillomaviruses (hrHPVs) infect mucosal epithelia and cause carcinomas at several anogenital sites, accounting for almost all cervical cancer cases worldwide.1 HPV is also a strong independent risk factor for oropharyngeal squamous cell carcinoma (OPSCC), a subset of head and neck cancers that occur in the epithelia lining lymphoid crypts of the tonsils and tongue base.2 Head and neck squamous cell carcinomas (HNSCCs, including those OPSCCs unrelated to HPV) typically occur in patients older than 60 years with a history of heavy smoking and high alcohol intake; HPV-driven OPSCCs affect a younger population, with a median age at diagnosis of younger than 60 years and often with little or no smoking history.3
Rates of HPV-negative (HPV−) HNSCC have begun to decline in the United States, Europe, and Australia; by contrast, HPV-driven OPSCC incidence is rapidly increasing in these countries.4 Although prophylactic vaccination against hrHPV types has the potential to prevent HPV-driven OPSCC, the sustained lifetime risk of oral hrHPV infection, coupled with a later age at diagnosis than cervical cancer, has led to the prediction that it will be at least 2060 before the current rising trend is reversed, emphasizing the need to correctly diagnose and treat these tumors.3 The clinical behavior of HPV-driven OPSCC also differs from HPV− disease, having significantly better progression-free and overall survival.5,6 These observations have led to HPV status becoming a routine molecular test for patients with OPSCC, and also have resulted in clinical trials examining de-intensification of treatment of these patients in an attempt to reduce the morbidity associated with the aggressive chemoradiotherapy regimens used for HNSCC.7
hrHPV DNA has been detected in tumors from other head and neck sites (non-OPSCCs) at varying frequencies of 5% to 15%, raising an important question as to whether these tumors are truly HPV driven and, if so, what implications this may have for their clinical behavior and management.8-11 Previous studies in this area have been limited by the fact that HPV DNA detection is not a very reliable marker of HPV causation, whereas detection of the p16INK4A tumor suppressor protein by immunohistochemistry (a clinical biomarker for HPV+ OPSCC) seems much less specific outside of the oropharynx; thus the significance of HPV detection in non-OPSCC remains unclear.8,10
In OPSCC, detection of transcripts encoding the E6 and E7 viral oncogenes is the gold standard for HPV causation and characteristic molecular profiles have been identified for these tumors.12,13 We investigated the role of HPV in non-OPSCC by examining molecular profiles of these tumors and comparing them to profiles we established from meta-analyses of known HPV-driven tumors and tumor models. Using machine learning, we generated gene expression and DNA methylation-based classifiers to predict the HPV status of 520 RNA-sequenced and 464 methylation-profiled HNSCCs that have now been profiled by The Cancer Genome Atlas (TCGA) project, including 75 HPV transcript-positive tumors (54 OPSCC, 21 non-OPSCC).14 We also evaluated the mutational spectra, genomic alterations, and morphologic characteristics previously shown to characterize HPV-driven HNSCC. Finally, we investigated the prognostic significance of HPV involvement outside of the oropharynx and related this to differences in the immune microenvironment between HPV-driven HNSCCs from different sites.
METHODS
Gene expression microarray data were downloaded from Array Express15 (www.ebi.ac.uk/arrayexpress). TCGA HNSCC molecular data and clinical meta-data were obtained from TCGA Data Portal (https://tcga-data.nci.nih.gov/tcga/) and the University of California, Santa Cruz, Cancer Browser (https://genome-cancer.ucsc.edu/), respectively. HPV transcript detection and quantification were conducted as described previously.16 Data processing and analysis were conducted using open-source tools in the R programming environment. The complete code used for all analyses is available from the authors upon request. High-resolution images from hematoxylin and eosin (H&E) –stained slides (TCGA digital slide archive, http://cancer.digitalslidearchive.net/) were scored blinded to anatomic site for tumor grade and tumor-infiltrating lymphocyte (TIL) content by a pathologist (G.J.T.), as described previously.17 For detailed descriptions of all methods, see the Data Supplement.
RESULTS
Meta-Analysis Defines a Cross-Tissue Transcriptional Signature for HPV-Driven Carcinogenesis
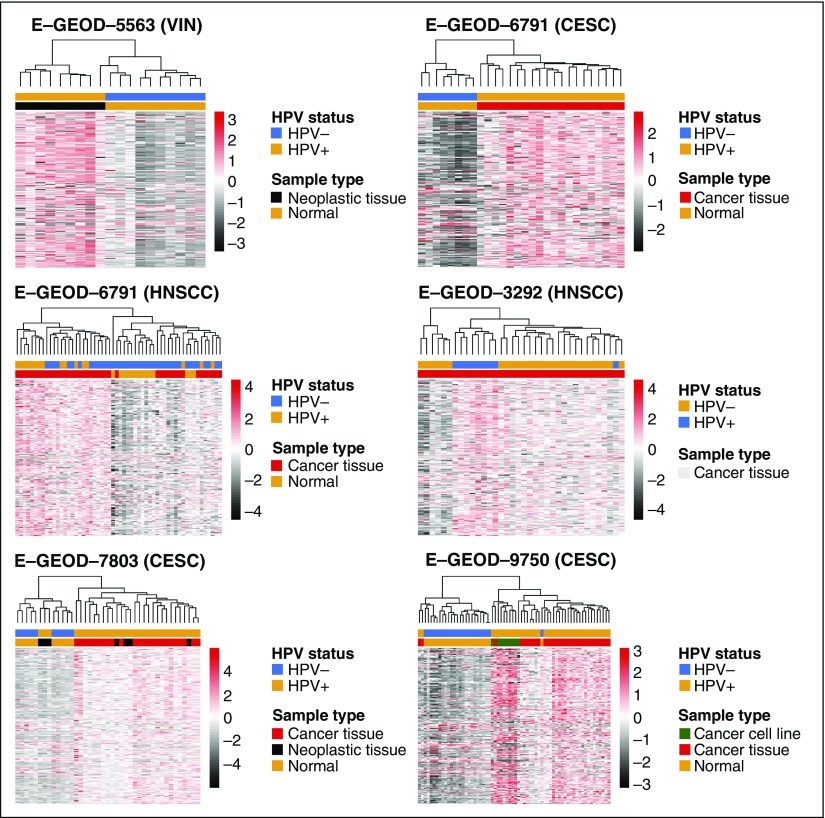
We conducted a meta-analysis of seven gene expression microarray (U133A, U133+2) studies that included tissue samples and cell-line models representing normal epithelium (n = 69), HPV− (n = 55), and HPV-driven (n = 138) tumors18-24 (Data Supplement Table A1). Effect size combination was used to account for cross-study/cross-platform batch effects.
A total of 159 genes were differentially expressed in the HPV-driven samples (false discovery rate [FDR] < 0.01; greater than two-fold change; Data Supplement Table A2). Differentially expressed gene (DEG) signature members showed a consistent pattern of deregulation across the datasets, as visualized by clustering (Clustering Index: range, 0% to 4.2%; Fig 1; Data Supplement Methods). Signature gene expression was also stronger in late passage/transformed HPV+ keratinocytes than in cells profiled immediately after HPV16 infection19 (Data Supplement Fig A1A). Furthermore, analysis of a gene expression dataset from mesenchymal stem cells transduced with E6 and/or E725 demonstrated that the signature genes were modulated by HPV oncogenes not only in a cross-tissue fashion but also in different cellular lineages (Data Supplement Fig A1B).
Fig 1.
Human papillomavirus (HPV)-driven tumors display a common gene expression signature. Meta-analysis identified 179 probes mapping to 159 gene probes as being significantly differentially expressed between HPV+ tumors and HPV− controls (tumor or normal). Annotations represent sample type data where relevant, and HPV status. CESC, cervical squamous cell carcinoma; HNSCC, squamous cell carcinoma of the head and neck; VIN, vulval intraepithelial neoplasia.
The 159 DEG Signature Serves as a Functional Readout for HPV Oncogene Activity
Consistent with published gene expression signatures for HPV-associated cancers,18,20,26-29 placing our DEG signature genes in a functional context using Ingenuity Pathway Analysis (Qiagen, Redwood City, CA) revealed expression changes consistent with remodeling of cell cycle progression by E7; notably CDKN2A and CCNE overexpression and CCND1 downregulation (Data Supplement Fig A2). Upstream regulatory analysis indicated inhibition of p53 and activation of E2F, again consistent with the well-characterized actions of the HPV oncoproteins on these pathways30 (Data Supplement Table A3). The 159-DEG signature also contains multiple genes previously found to be dysregulated in HPV-associated cancers, such as the meiotic synaptonemal complex component SYCP2, members of the replication licensing complex and kallikrein genes.13 Taken together, the expression signature reflects a causal role for HPV in these tumors.
HPV Transcript-Positive OPSCC and Non-OPSCC Share Common Gene Expression Patterns
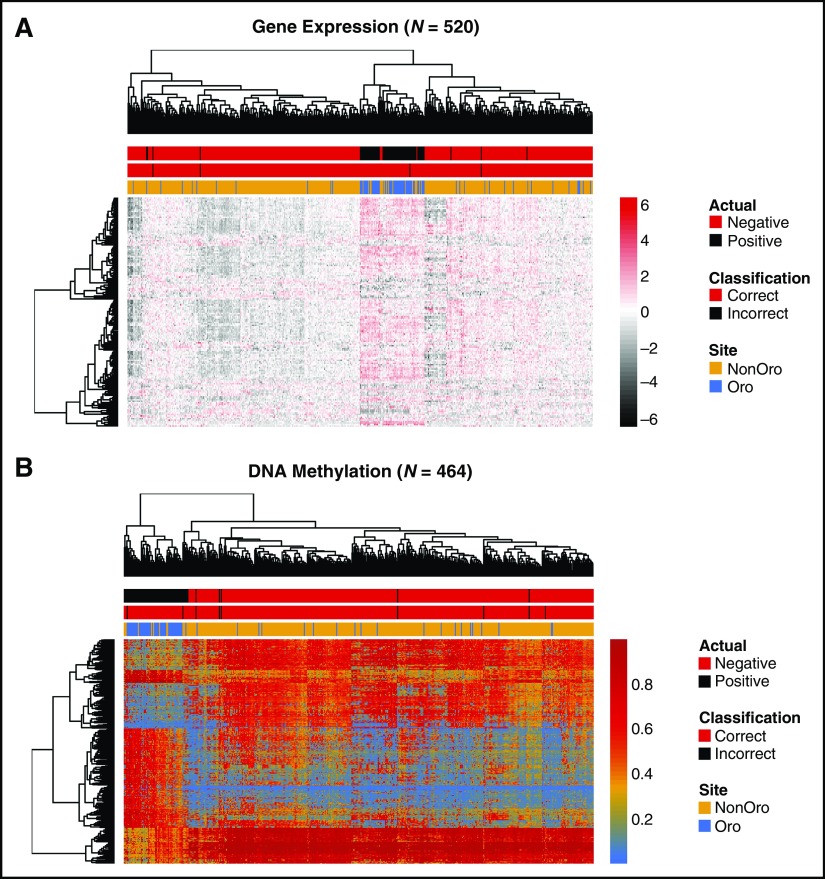
Next, we analyzed TCGA HNSC dataset,31 for which RNA-sequencing data are now available from 520 samples.14 We detected HPV transcripts in 54 of 80 OPSCC samples (67.5%) and 21 of 440 non-OPSCCs (4.8%); these are termed HPV+ (Data Supplement Table A4). Of the 159 expression signature genes, 127 were differentially expressed between HPV+ and HPV− samples (FDR < 0.01) in this data set. Almost all HPV+ samples (69 of 75) clustered together regardless of anatomic subsite, with OPSCCs and non-OPSCCs interspersed (Fig 2A).
Fig 2.
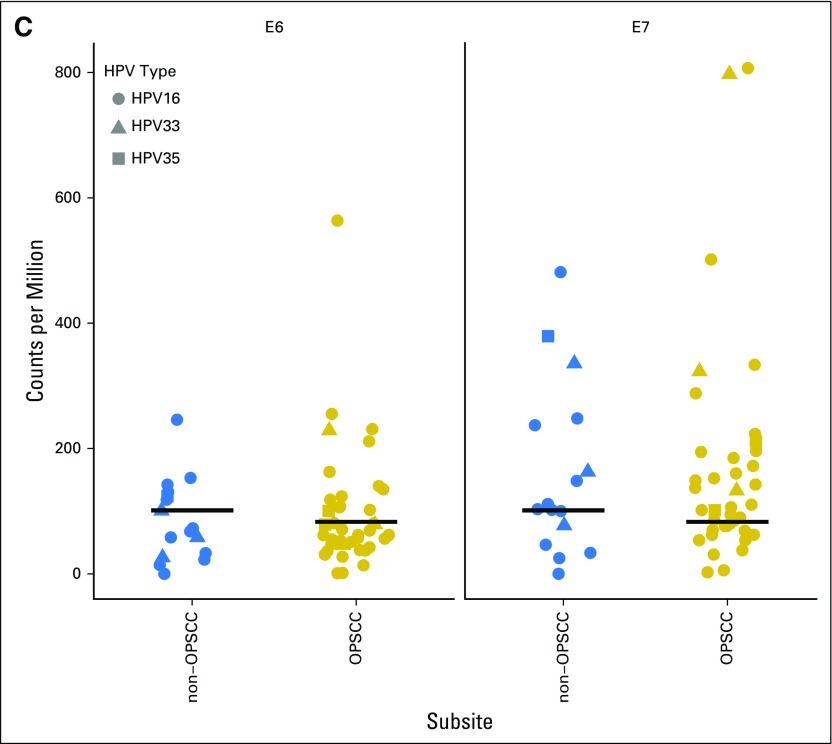
Molecular similarities between HPV transcript-positive tumors from different anatomic subsites. Heat maps show clustering of The Cancer Genome Atlas head and neck squamous cell carcinoma tumors using either (A) gene expression data (RNA-seq, n = 520; Illumina, San Diego, CA), on the basis of our 159-gene HPV signature or (B) DNA methylation data (Illumina 450k array, n = 464), on the basis of our 468 methylation variable position signature. Annotation bars indicate HPV status by RNA-seq analysis, OPSCC/non-OPSCC location, and predicted HPV status using our Random Forests classifier. HPV+ tumors generally clustered together, with non-OPSCC HPV+ tumors distributed among HPV+ OPSCCs. The gene expression heat map represents z-scores and the DNA methylation heat map displays β values. (C) Comparison of reads from the bicistronic HPV E6/E7 transcripts in OPSCC and non-OPSCC cases. HPV, human papillomavirus; nonOro, nonoropharyngeal head and neck cancer; OPSCC, oropharyngeal squamous cell carcinoma; Oro, oropharyngeal.
To test whether this gene set can accurately predict HPV status, we used two machine-learning approaches (the k-Nearest Neighbors [k-NN] algorithm and Random Forests [RF]; Salford Systems, San Diego, CA) to build classifiers. Both models performed well, returning out-of-fold kappa values (a measure of classifier accuracy) of 0.96 by RF and 0.94 by k-NN. The RF model correctly predicted all 445 HPV− and misclassified four of 75 HPV+ tumors (three non-OPSCC, one OPSCC), whereas the k-NN model misclassified four HPV+ and three HPV− tumors. Thus, the HPV status of OPSCCs and non-OPSCCs can be predicted using classifiers based upon our 159-DEG signature.
HPV Transcript-Positive OPSCC and Non-OPSCC Display Similar Genome-Wide DNA Methylation Landscapes
In addition to characteristic gene expression changes, several studies have reported distinct DNA methylation profiles between HPV+ and HPV− OPSCC, with HPV+ OPSCCs more closely resembling HPV+ cervical cancers than either HPV− OPSCC or lung squamous carcinoma.31-33 Using genome-wide DNA methylation profiles for only TCGA OPSCC samples (50 HPV+, 20 HPV−), we defined a 468-methylation variable position (MVP) signature (change in β [delta beta; dB] ≥ 0.4; FDR < 0.001; Data Supplement Table A5). When applied to the entire data set, this MVP signature clustered 60 of 69 HPV+ HNSCCs together, regardless of anatomic subsite (Fig 2B) and classifiers built using k-NN (κ = 0.93; none of 395 HPV− and seven of 69 HPV+ tumors misclassified) or RF (κ = 0.92; two of 395 HPV− samples, seven of 69 HPV+ samples misclassified) were able to predict the HPV status of non-OPSCCs as well as OPSCCs. These results support the conclusions, on the basis of gene expression data, that HPV+ OPSCCs and non-OPSCCs are molecularly similar and distinguishable from HPV− HNSCCs. These similarities in transcriptome and methylome profiles are further evident when visualized on multidimensional scaling plots (Data Supplement Fig A3). No MVPs differed between HPV+ OPSCC and HPV+ non-OPSCC at an FDR of 0.01/dB > 0.4, and only 19 differed at dB > 0.3.
HPV Oncogene Transcript Levels Do Not Vary Between HPV+ OPSCC and Non-OPSCC
Expression of the E6 and E7 viral oncogenes increases during HPV-driven tumor development and cells from these tumors are dependent on their continued expression. Assuming that if the HPV+ non-OPSCCs are, indeed, HPV-driven, they should express comparable E6/E7 levels as the HPV+ OPSCCs. We compared the levels of these transcripts and observed no difference between the two groups (Fig 2C). HPV+ non-OPSCCs, therefore, clearly express levels of E6 and E7 sufficient for the growth and survival of known HPV-driven tumors.
Genomic Similarities Between HPV+ OPSCC and Non-OPSCC
Having observed strong similarities between HPV+ OPSCC and non-OPSCC at the gene expression and epigenetic levels, we next examined the genomic features of these tumors. We previously reported elevated fractions of mutations attributable to deoxycytidine deaminase (APOBEC) activity in HPV+ HNSCC.34 Furthermore, APOBEC3B, previously implicated in this mutational process,35 was part of our gene expression signature (Data Supplement Table A2). The larger set of 502 HNSCCs that have now been exome sequenced by TCGA (including 68 HPV+) allowed us to compare enrichment for APOBEC signature mutations (C>T or C>G mutations occurring within the TCW trinucleotide motif, where W = A or T) in OPSCC versus non-OPSCC (Data Supplement Fig A4). Controlling for smoking history (which defines a separate subgroup within HPV+ OPSCC with respect to survival and APOBEC signature mutations5,36,37), HPV status remains a strong predictor for APOBEC independent of anatomic site (odds ratio, 3.07; P < 2.2 × 10−16), with HPV+ non-OPSCCs displaying an even higher proportion of APOBEC signature mutations than HPV+ OPSCC (odds ratio, 1.29; P = 2.2 × 10−9), again consistent with an HPV-driven etiology.
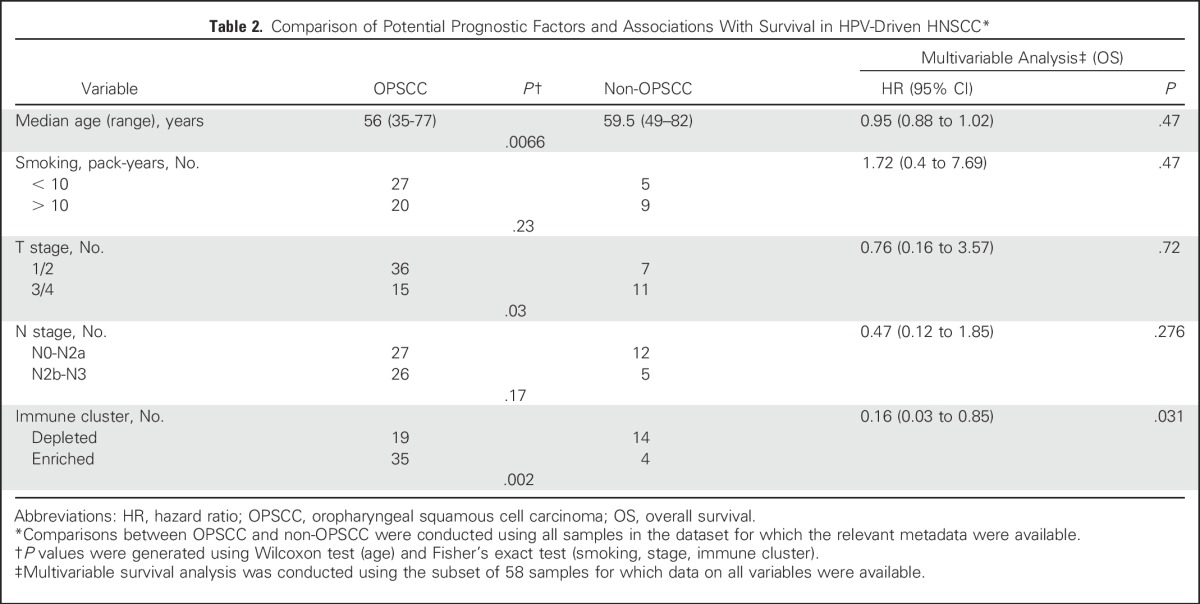
In addition to differences in global mutational signatures, HPV− and HPV+ OPSCCs display characteristic mutations and/or copy number alterations. TP53, CDKN2A, and CCND1 alterations are almost exclusive to HPV− OPSCCs, in which they occur at high frequency.31,38-41 In HPV+ OPSCCs, E6-mediated p53 degradation removes selection pressure for TP53 mutations, and bypass of the G1/S checkpoint by E7 likewise obviates pressure to acquire CDKN2A or CCND1 alterations.40,42 As expected, we observed high frequencies of TP53 mutations, CDKN2A mutations, deletions, and CCND1 amplifications in HPV− HNSCCs, and very low frequencies of these alterations in HPV+ OPSCC. Again, HPV+ non-OPSCC tumors closely resembled HPV+ OPSCCs (Table 1). Taken together, our molecular analyses strongly implicated HPV as a driver in 72 of 75 transcript-positive HNSCCs, and subsequent analyses focused on these tumors, hereafter termed HPV-driven (Data Supplement Methods).
Table 1.
Hallmark Genomic Alterations by Anatomic Site and HPV Status*
HPV-Driven Non-OPSCCs Frequently Display the Basaloid Morphology Typical of HPV-Driven OPSCCs
To complement our molecular observations, we conducted histopathological analysis, grading those TCGA cases for which evaluable images of H&E-stained sections were available through TCGA Digital Image Archive. As expected, the majority (39 of 41) HPV+ OPSCC cases displayed a characteristic basaloid (poorly differentiated) morphology, which has previously been linked to their origin in nonkeratinizing tonsillar crypt epithelium.43 Strikingly, whereas HNSCCs arising at other sites are typically keratinizing (well to moderately differentiated), 11 of the evaluable 15 HPV+ non-OPSCCs also displayed the basaloid morphology characteristic of the OPSCCs (Data Supplement Table A6). Consistent with these observations, comparison of global cytokeratin gene expression profiles revealed an intermediate pattern in the HPV-driven non-OPSCCs, suggesting they are determined by a combination of both HPV and anatomic site (Data Supplement Fig A5).
HPV-Driven Tumors Show Different Prognosis by Anatomic Subsite
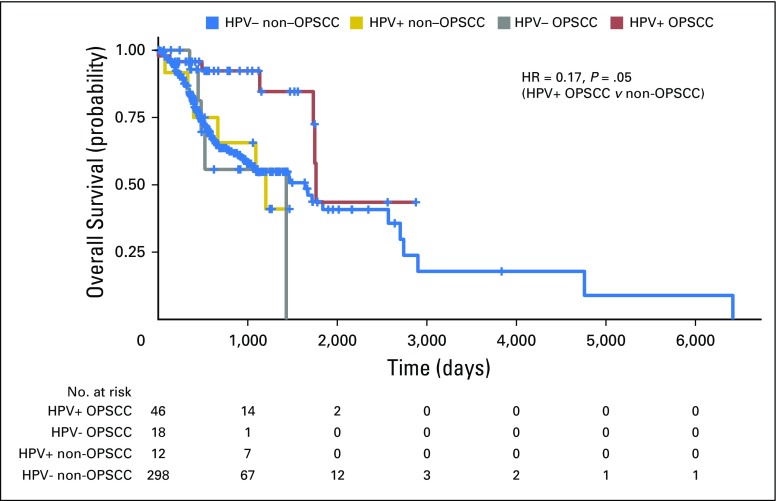
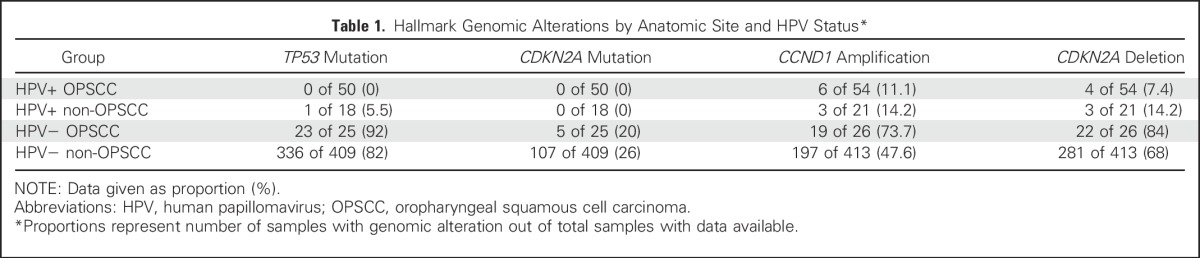
Patients with OPSCC with an HPV etiology show significantly improved survival compared with those with HPV− disease.5,6 Having demonstrated that HPV plays a causative role in transcript-positive non-OPSCC, we examined whether patients with these tumors display a similarly favorable prognosis. Kaplan-Meier analysis of TCGA cohort stratified into four groups by tumor HPV status and anatomic subsite suggested improved overall survival (OS) specifically in HPV-driven OPSCC (Fig 3). Accordingly, oropharyngeal location was associated with longer OS in HPV-driven tumors in multivariable analysis controlling for variables (ie, age, smoking, T stage, and N stage) previously associated with prognosis in HPV-driven disease.5,17
Fig 3.
Differences in overall survival by HPV status and anatomic subsite. Kaplan-Meier curves display overall survival in The Cancer Genome Atlas cohort for OPSCC (n = 77) and non-OPSCC (n = 432) by HPV status. The table indicates the number of patients still at risk by year. Statistics are from multivariable Cox regression controlling for age, smoking history (more or fewer than 10 pack-years), T stage (T1/2 v T3/4) and N stage (N0-N2a v N2b-N3). HPV, human papillomavirus; HR, hazard ratio; OPSCC, oropharyngeal squamous cell carcinoma.
Anatomic Subsite Is Associated With Differences in Lymphocyte Infiltration and Activation
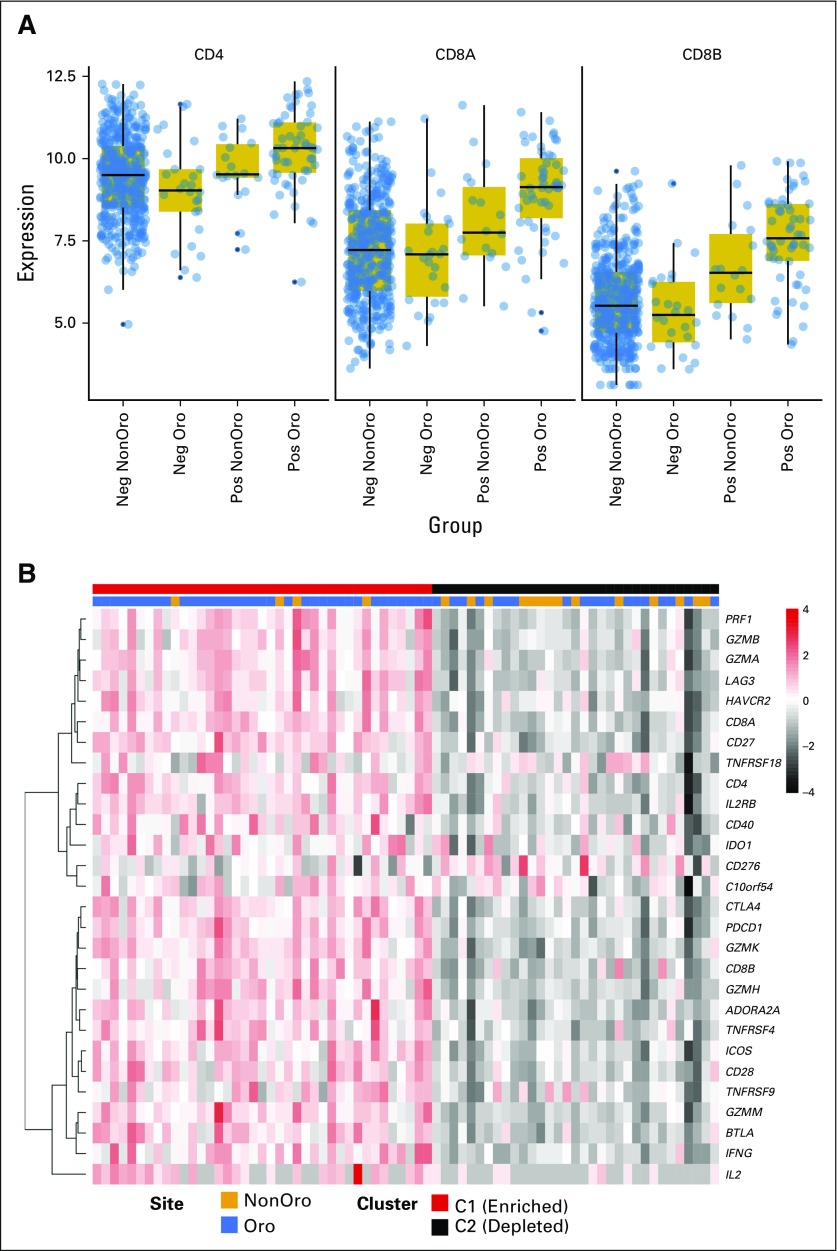
In addition to age, stage, and smoking history, TIL levels are reported to be associated with survival in HPV-driven OPSCC, with low TIL numbers identifying a subgroup of patients with poor prognosis and similar survival rates to those with HPV− OPSCCs.17 The importance of the immune system in shaping the evolution of HPV-driven OPSCC is also reflected by the high levels of programmed death ligand 1 expressed by these tumors.44 Therefore, we compared TIL levels between HPV-driven OPSCC and non-OPSCC by measuring CD4 (helper/regulatory T lymphocyte) and CD8A/CD8B (cytotoxic T lymphocyte) mRNA abundance; we found significantly higher levels of both mRNAs in OPSCC (Fig 4A). Consistent with this, we also observed increased TILs in H&E-stained sections17 from HPV-driven OPSCCs (Cochran-Armitage trend test, P = .046; Data Supplement Table A6) and found increased expression of multiple T-cell effector markers in HPV-driven OPSCC (Data Supplement Fig A6).
Fig 4.
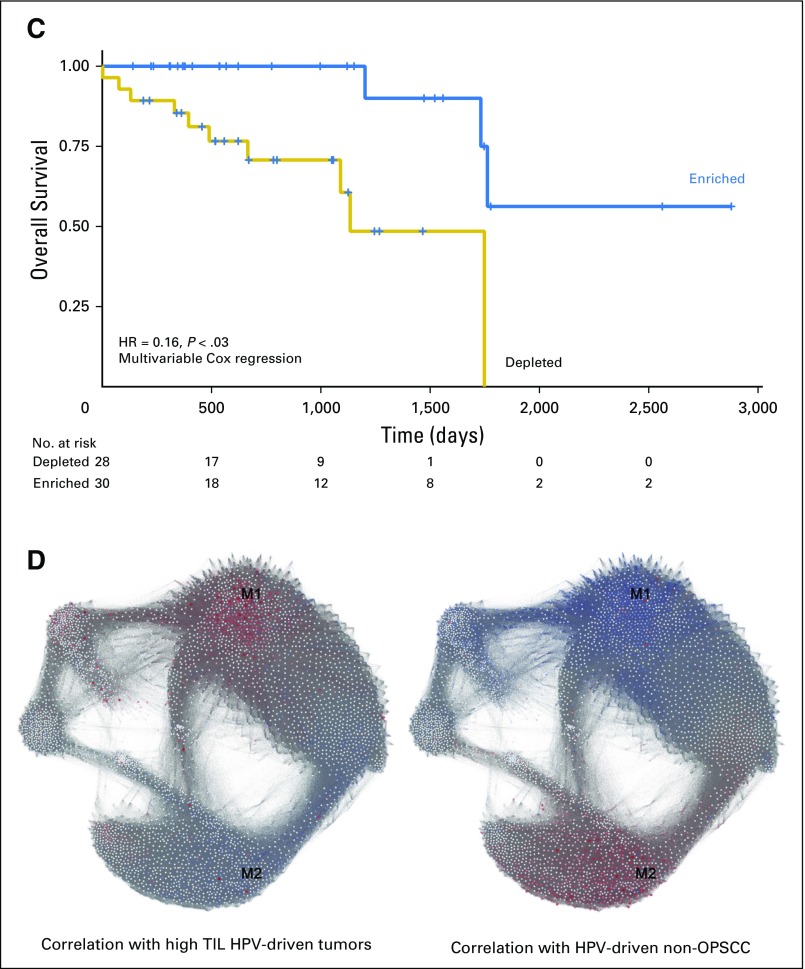
Enhanced lymphocyte infiltration in HPV-driven OPSCC. (A) Variance stabilization transformed mRNA levels of TIL markers CD4, CD8A, and CD8B. Each was significant at a false discover rate < 0.05 (Wilcoxon’s rank-sum test) comparing HPV-driven OPSCC with HPV-driven non-OPSCC. (B) The expression heat map of immune checkpoint gene transcripts highlights a low expression and a moderate/high expression cluster within HPV-driven squamous carcinoma of the head and neck (HNSCC); annotation bars represent anatomic subsite and cluster allocation from consensus clustering. (C) Kaplan-Meier curve of HPV+ HNSCC stratified by TIL status. The table represents the number at risk at the given time points. Statistics are from multivariable Cox regression controlling for age, smoking history (more or fewer than 10 pack-years), subsite, T stage (T1/2 v T3/4) and N stage (N0-N2a v N2b-N3). This also is compared in Table 2. (D) Weighted correlation network analysis network graphs. Each gene is labeled by color according to correlation with the indicated tumor subtype. Red and blue show positive and negative correlation, respectively. HPV, human papillomavirus; Neg, negative; OPSCC: oropharyngeal squamous cell carcinoma; Oro, oropharyngeal; Pos, positive; TIL, tumor-infiltrating lymphocyte.
Given the importance of immune checkpoints45,46 in modulating antitumor immune responses and the observation that PD1-expressing TILs are associated with favorable prognosis in HPV-driven OPSCC,47 we next investigated the clinical impact of TIL levels and their effector and immune checkpoint activity. Clustering HPV-driven tumors on the basis of their expression of a panel of T-cell genes denoting lineage, activation, and immune checkpoint expression revealed two clusters (Fig 4B). The low expression (immune-depleted) cluster was strongly enriched for non-OPSCCs (14 of 18 non-OPSCCs and 18 of 54 OPSCCs), with immune enrichment associated with longer OS in multivariable analysis (Fig 4C; Table 2). Finally, we examined relationships between the HPV-driven tumors and a previously published gene co-expression module (M1) enriched for genes associated with CD8+ T cells and improved survival in HNSCC (Fig 4D).48
Table 2.
Comparison of Potential Prognostic Factors and Associations With Survival in HPV-Driven HNSCC*
Gene expression in TIL-high HPV-driven tumors was correlated with this module (r = 0.46; 95% CI, 0.44 to 0.49; P < 2.2 × 10−16). HPV-driven non-OPSCCs, however, were inversely correlated with M1 (r = −0.65; 95% CI, −0.67 to −0.63; P < 2.2 × 10−16) and were, instead, mostly correlated (r = 0.66; 95% CI, 0.64 to 0.67; P < 2.2 × 10−16) with a module (M2) associated with upregulated glucose metabolism and poor outcome, overrepresented in tumors of the basal subtype not previously linked to HPV-driven HNSCC.48-50
Overall, the HPV-driven OPSCCs and non-OPSCCs were very similar; we found only 67 DEGs (Data Supplement Table A7) and no significant MVPs. However, taken together, our analyses reveal a clear, survival-associated difference in lymphocyte infiltration between HPV-driven OPSCC and non-OPSCC.
DISCUSSION
The importance of HPV in OPSCC and its implications for patient treatment and the design of clinical trials are well established. HPV is detected at lower frequencies in non-OPSCC, where its biologic role and clinical relevance are unclear. We hypothesized that if HPV were playing a driver role in these tumors, they would share molecular profiles of known HPV-driven cancers. Our study shows HPV+ non-OPSCCs do, indeed, share a gene expression signature consistent with an HPV-driven etiology. Furthermore, HPV+ HNSCCs are similar (and distinct from HPV− HNSCCs) at the epigenetic and genomic levels. HPV+ non-OPSCCs also frequently display the basaloid morphology that characterizes HPV+ OPSCC. This finding, also supported by observations from HPV-associated anogenital and sinonasal cancers, is consistent with a role for HPV in blocking the differentiation of cells that would normally give rise to keratinizing epithelia.51,52
Our findings suggest that viral etiology alone is insufficient to confer favorable prognosis in HNSCC and that this effect is largely limited to the oropharnynx; among non-OPSCC cases, any survival benefit of HPV seems minimal (hazard ratio, 0.82; 95% CI, 0.38 to 1.76; P = .63). These findings are supported by two previous studies, in which p16 expression in non-OPSCC was associated with either a more limited or no improvement in OS.8,10 We acknowledge that given the possible heterogeneity between tumors from different anatomic head and neck subsites, it is not ideal to class “non-OPSCC” as a single group—a fact also accepted by Chung and colleagues8 as a limitation in their prognostic analysis of p16 expression in non-OPSCC. We also recognize that although we see a clear difference in OS and lymphocyte infiltration and activity between HPV-driven OPSCC and non-OPSCC, there were insufficient data on treatment in this cohort to permit its inclusion as a variable in multivariable analysis. Investigation of a larger, uniformly treated patient cohort is needed, therefore, to confirm that the difference in prognosis is independent of possible differences in patient management between OPSCC and non-OPSCC. Notably, in OPSCC, the improved prognosis of patients with HPV-driven disease has been shown in several studies to be independent of treatment modality,53-55 arguing for a tumor-intrinsic factor in driving this survival difference. It has been suggested that this is due, at least in part, to increased radiosensitivity, perhaps resulting from retention of wild-type TP53.56 The almost universal retention of wild-type TP53 in HPV-driven HNSCC (Table 1) suggests an alternative explanation for the survival difference by subsite.
The difference in TIL levels between HPV-driven OPSCC and non-OPSCC suggests a fundamentally different immune response to these tumors. Interestingly, in their analysis of the initial HNSCC dataset of 279 tumors, 14 of which were HPV transcript-positive non-OPSCCs, TCGA classified several as belonging to the basal transcriptional subtype initially identified by Chung and colleagues.31,49 Our analysis demonstrates that these tumors are, in fact, highly similar to HPV-driven OPSCCs of the atypical subtype, and that they are also HPV-driven but lack the lymphocyte infiltration associated with improved outcomes in OPSCC. This observation potentially explains the survival differences between HPV-driven tumors at these sites and implies that there is something unique to the oropharynx that enhances the immune response to HPV-driven tumors (but not HPV− tumors) arising at this location.
Our analysis identifies and provides important insight into a new patient population, with implications for both cancer prevention and treatment. Using multiomic TCGA data has uniquely allowed us to build a comprehensive molecular map of HPV+ HNSCCs and to use HPV transcript detection as our marker for HPV positivity throughout, thus identifying a significant role for HPV outside the oropharynx and avoiding the previously documented pitfalls of either HPV DNA or p16 detection.8,57 HPV transcript-positive non-OPSCCs are HPV driven and would likely benefit from HPV-targeted therapies (and be prevented by prophylactic HPV vaccination). The relatively poor prognosis of HPV-driven non-OPSCC, however, argues against treatment de-escalation for these tumors, whereas clear differences in the immune microenvironment suggest we may expect altered responses to immune checkpoint blockade. From a precision medicine perspective, HPV-driven non-OPSCC cases should be identified and considered as a distinct entity with respect to both HPV− HNSCC and HPV-driven OPSCC.
ACKNOWLEDGMENT
The results published here are, in part, based upon data generated by The Cancer Genome Atlas (TCGA) project established by the National Cancer Institute and National Human Genome Research Institute. Information about TCGA and the investigators and institutions who constitute TCGA research network can be found at http://cancergenome.nih.gov.
Footnotes
Supported by grants from Rosetrees Trust (T.R.F.) and Cancer Research UK (S.H., G.J.T.). A.F. is supported by the MRC (MR/M025411/1) and UCLH/UCL Comprehensive Biomedical Research Programme. A.C. is supported by a graduate scholarship from University College London.
Presented at the annual meeting of the American Association for Cancer Research, New Orleans, LA, April 16-20, 2016, and at the Midwest Epigenetics and Chromatin Meeting, Grand Rapids, MI, June 5-7, 2016.
Authors’ disclosures of potential conflicts of interest are found in the article online at www.jco.org. Author contributions are found at the end of this article.
AUTHOR CONTRIBUTIONS
Conception and design: Ankur Chakravarthy, Stephen Henderson, Stephen M. Thirdborough, Christian H. Ottensmeier, Andrew Feber, Gareth J. Thomas, Tim R. Fenton
Collection and assembly of data: Ankur Chakravarthy, Stephen Henderson, Xiaoping Su, Tim R. Fenton
Data analysis and interpretation: Ankur Chakravarthy, Stephen Henderson, Stephen M. Thirdborough, Christian H. Ottensmeier, Andrew Feber, Matt Lechner, Gareth J. Thomas, Tim R. Fenton
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
HPV Drives Tumor Development Throughout the Head and Neck: Improved Prognosis Is Associated With an Immune Response Largely Restricted to the Oropharynx
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or jco.ascopubs.org/site/ifc.
Ankur Chakravarthy
No relationship to disclose
Stephen Henderson
No relationship to disclose
Stephen M. Thirdborough
No relationship to disclose
Christian H. Ottensmeier
Speakers' Bureau: Bristol-Myers Squibb, Roche, Delcath Systems
Research Funding: Bristol-Myers Squibb (Inst), Amgen (Inst), Asterias (Inst), BioNTech AG (Inst), MedImmune (Inst), MSD (Inst), Touchlight Genetics (Inst), Verastem (Inst)
Xiaoping Su
No relationship to disclose
Matt Lechner
No relationship to disclose
Andrew Feber
No relationship to disclose
Gareth J. Thomas
Consulting or Advisory Role: GlaxoSmithKline, MSD
Speakers' Bureau: Almiral
Tim R. Fenton
No relationship to disclose
REFERENCES
- 1.Doorbar J, Quint W, Banks L, et al. The biology and life-cycle of human papillomaviruses. Vaccine. 2012;30(suppl 5):F55–F70. doi: 10.1016/j.vaccine.2012.06.083. [DOI] [PubMed] [Google Scholar]
- 2.Goon PK, Stanley MA, Ebmeyer J, et al. HPV & head and neck cancer: A descriptive update. Head Neck Oncol. 2009;1:36. doi: 10.1186/1758-3284-1-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gillison ML, Chaturvedi AK, Anderson WF, et al. Epidemiology of human papillomavirus-positive head and neck squamous cell carcinoma. J Clin Oncol. 2015;33:3235–3242. doi: 10.1200/JCO.2015.61.6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaturvedi AK, Engels EA, Pfeiffer RM, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29:4294–4301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fakhry C, Westra WH, Li S, et al. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100:261–269. doi: 10.1093/jnci/djn011. [DOI] [PubMed] [Google Scholar]
- 7.Vokes EE, Agrawal N, Seiwert TY. HPV-associated head and neck cancer. J Natl Cancer Inst. 2015;107:djv344. doi: 10.1093/jnci/djv344. [DOI] [PubMed] [Google Scholar]
- 8.Chung CH, Zhang Q, Kong CS, et al. p16 protein expression and human papillomavirus status as prognostic biomarkers of nonoropharyngeal head and neck squamous cell carcinoma. J Clin Oncol. 2014;32:3930–3938. doi: 10.1200/JCO.2013.54.5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herrero R, Castellsagué X, Pawlita M, et al. Human papillomavirus and oral cancer: The International Agency for Research on Cancer multicenter study. J Natl Cancer Inst. 2003;95:1772–1783. doi: 10.1093/jnci/djg107. [DOI] [PubMed] [Google Scholar]
- 10.Lassen P, Primdahl H, Johansen J, et al. Impact of HPV-associated p16-expression on radiotherapy outcome in advanced oropharynx and non-oropharynx cancer. Radiother Oncol. 2014;113:310–316. doi: 10.1016/j.radonc.2014.11.032. [DOI] [PubMed] [Google Scholar]
- 11.Walline HM, Komarck C, McHugh JB, et al. High-risk human papillomavirus detection in oropharyngeal, nasopharyngeal, and oral cavity cancers: Comparison of multiple methods. JAMA Otolaryngol Head Neck Surg. 2013;139:1320–1327. doi: 10.1001/jamaoto.2013.5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung CH, Gillison ML. Human papillomavirus in head and neck cancer: Its role in pathogenesis and clinical implications. Clin Cancer Res. 2009;15:6758–6762. doi: 10.1158/1078-0432.CCR-09-0784. [DOI] [PubMed] [Google Scholar]
- 13. Lechner M, Fenton TR: The genomics, epigenomics and transcriptomics of HPV-associated oropharyngeal cancer - understanding the basis of a rapidly evolving disease, in Friedmann T, Dunlap JC, Goodwin SF (eds): Advances in Genetics. Cambridge, MA, Academic Press, 2016. [DOI] [PubMed] [Google Scholar]
- 14. The Cancer Genome Atlas: TCGA Data Portal. https://tcga-data.nci.nih.gov/docs/publications/tcga/
- 15.Kolesnikov N, Hastings E, Keays M, et al. ArrayExpress update--Simplifying data submissions. Nucleic Acids Res. 2015;43:D1113–D1116. doi: 10.1093/nar/gku1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y, Yao H, Thompson EJ, et al. VirusSeq: Software to identify viruses and their integration sites using next-generation sequencing of human cancer tissue. Bioinformatics. 2013;29:266–267. doi: 10.1093/bioinformatics/bts665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ward MJ, Thirdborough SM, Mellows T, et al. Tumour-infiltrating lymphocytes predict for outcome in HPV-positive oropharyngeal cancer. Br J Cancer. 2014;110:489–500. doi: 10.1038/bjc.2013.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pyeon D, Newton MA, Lambert PF, et al. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res. 2007;67:4605–4619. doi: 10.1158/0008-5472.CAN-06-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kravchenko-Balasha N, Mizrachy-Schwartz S, Klein S, et al. Shift from apoptotic to necrotic cell death during human papillomavirus-induced transformation of keratinocytes. J Biol Chem. 2009;284:11717–11727. doi: 10.1074/jbc.M900217200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slebos RJC, Yi Y, Ely K, et al. Gene expression differences associated with human papillomavirus status in head and neck squamous cell carcinoma. Clin Cancer Res. 2006;12:701–709. doi: 10.1158/1078-0432.CCR-05-2017. [DOI] [PubMed] [Google Scholar]
- 21.Santegoets LA, Seters Mv, Helmerhorst TJ, et al. HPV related VIN: Highly proliferative and diminished responsiveness to extracellular signals. Int J Cancer. 2007;121:759–766. doi: 10.1002/ijc.22769. [DOI] [PubMed] [Google Scholar]
- 22.Scotto L, Narayan G, Nandula SV, et al. Identification of copy number gain and overexpressed genes on chromosome arm 20q by an integrative genomic approach in cervical cancer: Potential role in progression. Genes Chromosomes Cancer. 2008;47:755–765. doi: 10.1002/gcc.20577. [DOI] [PubMed] [Google Scholar]
- 23.Zhai Y, Kuick R, Nan B, et al. Gene expression analysis of preinvasive and invasive cervical squamous cell carcinomas identifies HOXC10 as a key mediator of invasion. Cancer Res. 2007;67:10163–10172. doi: 10.1158/0008-5472.CAN-07-2056. [DOI] [PubMed] [Google Scholar]
- 24. EMBL-EBI: E-GEOD-24089 - Expression data from HPV(-) and HPV(+) squamous cell carcinoma cell lines. 2010. http://www.ebi.ac.uk/arrayexpress/experiments/E-GEOD-24089/samples/ [Google Scholar]
- 25.Funes JM, Quintero M, Henderson S, et al. Transformation of human mesenchymal stem cells increases their dependency on oxidative phosphorylation for energy production. Proc Natl Acad Sci USA. 2007;104:6223–6228. doi: 10.1073/pnas.0700690104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung AC, Briolat J, Millon R, et al. Biological and clinical relevance of transcriptionally active human papillomavirus (HPV) infection in oropharynx squamous cell carcinoma. Int J Cancer. 2010;126:1882–1894. doi: 10.1002/ijc.24911. [DOI] [PubMed] [Google Scholar]
- 27.Lohavanichbutr P, Houck J, Fan W, et al. Genomewide gene expression profiles of HPV-positive and HPV-negative oropharyngeal cancer: Potential implications for treatment choices. Arch Otolaryngol Head Neck Surg. 2009;135:180–188. doi: 10.1001/archoto.2008.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlecht NF, Burk RD, Adrien L, et al. Gene expression profiles in HPV-infected head and neck cancer. J Pathol. 2007;213:283–293. doi: 10.1002/path.2227. [DOI] [PubMed] [Google Scholar]
- 29.Tang K-W, Alaei-Mahabadi B, Samuelsson T, et al. The landscape of viral expression and host gene fusion and adaptation in human cancer. Nat Commun. 2013;4:2513. doi: 10.1038/ncomms3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Münger K, Baldwin A, Edwards KM, et al. Mechanisms of human papillomavirus-induced oncogenesis. J Virol. 2004;78:11451–11460. doi: 10.1128/JVI.78.21.11451-11460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Network Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lechner M, Fenton T, West J, et al. Identification and functional validation of HPV-mediated hypermethylation in head and neck squamous cell carcinoma. Genome Med. 2013;5:15. doi: 10.1186/gm419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parfenov M, Pedamallu CS, Gehlenborg N, et al. Characterization of HPV and host genome interactions in primary head and neck cancers. Proc Natl Acad Sci USA. 2014;111:15544–15549. doi: 10.1073/pnas.1416074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henderson S, Chakravarthy A, Su X, et al. APOBEC-mediated cytosine deamination links PIK3CA helical domain mutations to human papillomavirus-driven tumor development. Cell Reports. 2014;7:1833–1841. doi: 10.1016/j.celrep.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 35.Burns MB, Lackey L, Carpenter MA, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. doi: 10.1128/mBio.02234-14. Vieira VC, Leonard B, White EA, et al: Human papillomavirus E6 triggers upregulation of the antiviral and cancer genomic DNA deaminase APOBEC3B. MBio 5(6) 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weinberger PM, Yu Z, Haffty BG, et al. Molecular classification identifies a subset of human papillomavirus-associated oropharyngeal cancers with favorable prognosis. J Clin Oncol. 2006;24:736–747. doi: 10.1200/JCO.2004.00.3335. [DOI] [PubMed] [Google Scholar]
- 38.Braakhuis BJM, Snijders PJF, Keune W-JH, et al. Genetic patterns in head and neck cancers that contain or lack transcriptionally active human papillomavirus. J Natl Cancer Inst. 2004;96:998–1006. doi: 10.1093/jnci/djh183. [DOI] [PubMed] [Google Scholar]
- 39.Gillison ML, Koch WM, Capone RB, et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92:709–720. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 40.Lechner M, Frampton GM, Fenton T, et al. Targeted next-generation sequencing of head and neck squamous cell carcinoma identifies novel genetic alterations in HPV+ and HPV- tumors. Genome Med. 2013;5:49. doi: 10.1186/gm453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Houten VMM, Snijders PJF, van den Brekel MWM, et al. Biological evidence that human papillomaviruses are etiologically involved in a subgroup of head and neck squamous cell carcinomas. Int J Cancer. 2001;93:232–235. doi: 10.1002/ijc.1313. [DOI] [PubMed] [Google Scholar]
- 42.Westra WH, Taube JM, Poeta ML, et al. Inverse relationship between human papillomavirus-16 infection and disruptive p53 gene mutations in squamous cell carcinoma of the head and neck. Clin Cancer Res. 2008;14:366–369. doi: 10.1158/1078-0432.CCR-07-1402. [DOI] [PubMed] [Google Scholar]
- 43.Shah KV, Westra WH. Genital HPVs in the aerodigestive tract: Etiologic association with a subset of oropharyngeal/tonsillar cancers and with recurrent respiratory papillomatosis. Dis Markers. 2007;23:235–245. doi: 10.1155/2007/913761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lyford-Pike S, Peng S, Young GD, et al. Evidence for a role of the PD-1:PD-L1 pathway in immune resistance of HPV-associated head and neck squamous cell carcinoma. Cancer Res. 2013;73:1733–1741. doi: 10.1158/0008-5472.CAN-12-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Medler TR, Cotechini T, Coussens LM. Immune response to cancer therapy: Mounting an effective antitumor response and mechanisms of resistance. Trends Cancer. 2015;1:66–75. doi: 10.1016/j.trecan.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Śledzińska A, Menger L, Bergerhoff K, et al. Negative immune checkpoints on T lymphocytes and their relevance to cancer immunotherapy. Mol Oncol. 2015;9:1936–1965. doi: 10.1016/j.molonc.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Badoual C, Hans S, Merillon N, et al. PD-1-expressing tumor-infiltrating T cells are a favorable prognostic biomarker in HPV-associated head and neck cancer. Cancer Res. 2013;73:128–138. doi: 10.1158/0008-5472.CAN-12-2606. [DOI] [PubMed] [Google Scholar]
- 48. Ottensmeier CH, Perry KL, Harden EL, et al: Upregulated glucose metabolism correlates inversely with CD8+ T-cell infiltration and survival in squamous cell carcinoma. Cancer Res (in press) [DOI] [PubMed]
- 49.Chung CH, Parker JS, Karaca G, et al. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell. 2004;5:489–500. doi: 10.1016/s1535-6108(04)00112-6. [DOI] [PubMed] [Google Scholar]
- 50.Keck MK, Zuo Z, Khattri A, et al. Integrative analysis of head and neck cancer identifies two biologically distinct HPV and three non-HPV subtypes. Clin Cancer Res. 2015;21:870–881. doi: 10.1158/1078-0432.CCR-14-2481. [DOI] [PubMed] [Google Scholar]
- 51.El-Mofty SK, Lu DW. Prevalence of high-risk human papillomavirus DNA in nonkeratinizing (cylindrical cell) carcinoma of the sinonasal tract: A distinct clinicopathologic and molecular disease entity. Am J Surg Pathol. 2005;29:1367–1372. doi: 10.1097/01.pas.0000173240.63073.fe. [DOI] [PubMed] [Google Scholar]
- 52.Westra WH. The morphologic profile of HPV-related head and neck squamous carcinoma: Implications for diagnosis, prognosis, and clinical management. Head Neck Pathol. 2012;6((suppl 1)):S48–S54. doi: 10.1007/s12105-012-0371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fischer CA, Zlobec I, Green E, et al. Is the improved prognosis of p16 positive oropharyngeal squamous cell carcinoma dependent of the treatment modality? Int J Cancer. 2010;126:1256–1262. doi: 10.1002/ijc.24842. [DOI] [PubMed] [Google Scholar]
- 54.Hong AM, Dobbins TA, Lee CS, et al. Human papillomavirus predicts outcome in oropharyngeal cancer in patients treated primarily with surgery or radiation therapy. Br J Cancer. 2010;103:1510–1517. doi: 10.1038/sj.bjc.6605944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ward MJ, Mellows T, Harris S, et al. Staging and treatment of oropharyngeal cancer in the human papillomavirus era. Head Neck. 2015;37:1002–1013. doi: 10.1002/hed.23697. [DOI] [PubMed] [Google Scholar]
- 56.Kimple RJ, Smith MA, Blitzer GC, et al. Enhanced radiation sensitivity in HPV-positive head and neck cancer. Cancer Res. 2013;73:4791–4800. doi: 10.1158/0008-5472.CAN-13-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seiwert TY. Ties that bind: p16 as a prognostic biomarker and the need for high-accuracy human papillomavirus testing. J Clin Oncol. 2014;32:3914–3916. doi: 10.1200/JCO.2014.57.9268. [DOI] [PubMed] [Google Scholar]