Abstract
BACKGROUND
In the past 3 decades, a better understanding of gene mutations and their role in carcinogenesis has led to improvement in our ability to treat patients with metastatic disease. The objective of the current study was to determine whether the location of a driver mutation within an affected gene impacts the biology of metastatic colorectal cancer.
METHODS
DNA was collected from 165 randomly selected specimens of patients who underwent margin-negative resection of colorectal liver metastases with curative intent. Sequenom analysis and Sanger sequencing were used to evaluate mutations in K/NRAS, PIK3CA, BRAF, and TP53.
RESULTS
BRAF mutation was associated with early recurrence and death, whereas no impact of TP53 or PIK3CA mutation was identified. Although K/NRAS mutation was associated with worse survival in this cohort, this difference was no longer evident when those who had received anti-EGFR therapy were excluded. When stratifying patients according to the exon on which K/NRAS was mutated, there were dramatic differences in both survival and pathologic features. Exon 4 mutations were associated with large, solitary metastases occurring at long disease-free intervals compared with exon 3 mutations, which presented with small, numerous lesions. Patients who had exon 4 mutations recurred infrequently and had significantly longer survival compared with those who had wild type or other mutations.
CONCLUSIONS
By using this model of curative-intent, margin-negative resection in patients at high risk of recurrence, the authors were able to establish a link between mutation location within the K/NRAS gene and the biology of metastatic colorectal cancer.
Keywords: colorectal metastases, hepatectomy, rat sarcoma (RAS) mutation, surgery, v-Raf murine sarcoma viral oncogene homolog B1 (BRAF) mutation
INTRODUCTION
Colorectal cancer represents the third most common malignancy in men and women in the United States with 135,000 new cases per year. Twenty percent of patients present with metastatic disease, and many more develop distant spread during the course of their illness.1 The most common site of metastasis is the liver: nearly 80% of patients who have stage IV disease have liver metastasis, and the liver is sole site of disease in 40%. Resection of colorectal liver metastases (CRLM) is associated with 5-year survival rates of 40% to 60%2,3 as well as occasional long-term cures. The 5-year survival rate with systemic chemotherapy alone for stage IV disease approaches 10% in highly selected patients, requires chronic therapy, and is typically noncurative.2,4,5 Although risk scores based on clinical and pathologic parameters have been developed to predict outcomes after hepatic metastasectomy, these prognostic models tend not to impact clinical decision making and do not translate well across institutions.6 Therefore, novel and effective prognostic biomarkers are of obvious importance. At the root of the problem is a lack of convincing data connecting the biologic drivers of carcinogenesis to the phenotype and survival after metastasectomy.
The mitogen-activated protein kinase (MAPK) signaling pathway consists of a series of kinases that, when triggered by an extracellular signal, results in a downstream cascade influencing cellular proliferation, differentiation, and survival. In the middle 1980s, it was discovered that mutations in MAPK pathway genes, most notably Kirstin rat sarcoma viral oncogene (K/NRAS), were integral to the initiation and progression of up to 50% of colorectal adenocarcinomas.7 Since then, oncogenic mutations in other genes, including v-Raf murine sarcoma viral oncogene (BRAF)8 and phosphoinositide 3-kinase catalytic subunit α (PIK3CA),9 have been identified. Therapies targeting these pathways have been developed, and RAS mutational status has emerged as a major predictor of response to anti-epidermal growth factor receptor (anti-EGFR) therapies.10,11
Controversy exists regarding the impact of MAPK mutations on recurrence and survival in patients with colorectal cancer. Previous studies investigating the association of MAPK mutations with the phenotype of metastases and impact on survival have been limited by 3 major factors: 1) the inclusion of patients with a wide variety of tumor burden and stage, 2) failure to account for differences because of receipt of anti-EGFR therapy, and 3) lack of analysis of less frequently mutated genes and exons, such as neuroblastoma rat sarcoma viral oncogene homolog (NRAS) and KRAS exons 3 and 4.
The objective of this study was to assess the impact of KRAS, NRAS, BRAF, PIK3CA, and tumor protein 53 (TP53) mutations on recurrence and survival in patients undergoing curative intent complete resection of CRLM. This population of patients is at high risk of recurrence, although they present without macroscopic disease, which reduces disease burden as a confounder.
MATERIALS AND METHODS
Patients
A prospectively maintained database of tissue samples obtained under an institutional review board-approved protocol was queried to identify patients who underwent resection of CRLM. All patients who had formalin-fixed, paraffin-embedded (FFPE) blocks and matched frozen tissue available from resected colorectal liver metastases were selected for further analysis. Perioperative and survival data were collected by review of a prospectively maintained clinical database and supplemented by a retrospective review of the medical record. Chemotherapy administration records were queried for administration of the anti-EGFR inhibitors cetuximab or panitumumab either before or after surgery. Only patients who underwent margin-negative (R0), curative-intent hepatic resection were included. Pathology reports were used to capture tumor size and number and to confirm margin status.
DNA Extraction and Sequencing
Hematoxylin and eosin-stained slides prepared from FFPE tissue were reviewed by a gastrointestinal pathologist (E.V.) to ensure adequate tumor content (>50%). Tumors with excessive necrosis were excluded from analysis. Genomic DNA was extracted from colorectal liver metastasis tissue using the QIAmp DNA extraction kit (Qiagen, Valencia, Calif) according to the manufacturer’s protocol and was subjected to whole genome amplification using the Repli-G Midi kit (Qiagen). The quality of whole genome-amplified DNA was verified by polymerase chain reactions using 2 control amplicons. The mass array-based iPLEX assay (Sequenom, San Diego, Calif) was used to detect mutations in KRAS (codons 12, 13, 22, 61, 117, and 146), NRAS (codons 12, 13, and 61), BRAF (codon 600), and PIK3CA (codons 345, 420, 542, 545, 546, 1043, and 1047), as previously described.12,13 All mutations were confirmed by repeat iPLEX assay or Sanger sequencing. TP53 mutations were assessed by routine Sanger sequencing.
Statistical Analysis
Nominal variables were evaluated using 2-tailed chi-square or Fisher exact tests, as appropriate. Continuous variables were assessed with univariate logistic regression for parametric values and the Wilcoxon rank-sum test for nonparametric values. Kaplan-Meier survival curves were created to determine differences in time to recurrence and survival. Multivariate regression was performed using data that trended toward significance (P <.10) on univariate analysis and variables that were previously associated with the outcome. P values <.05 were considered significant.
RESULTS
Patients
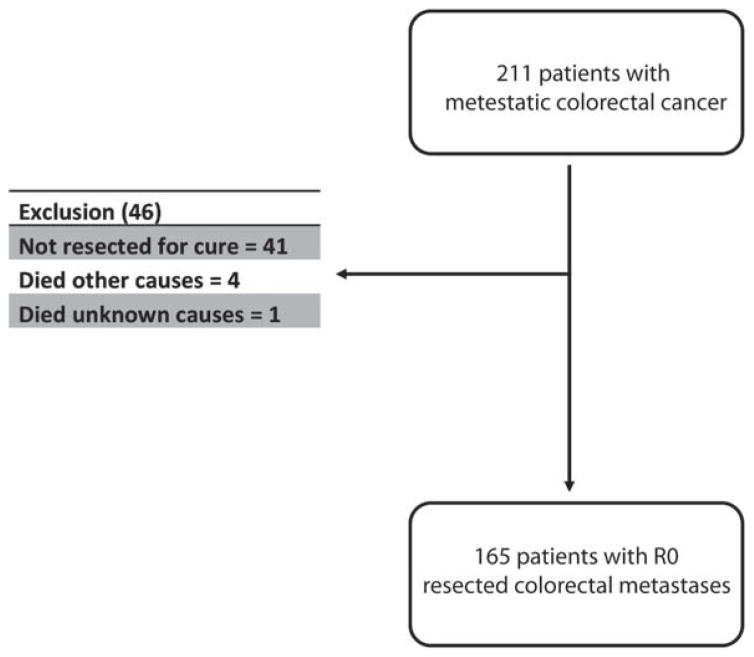
After a medical records review of patients who underwent resection for colorectal cancer metastases between 1992 and 2009, we identified 211 patients who underwent resection of CRLM and had FFPE and frozen tissue samples available for further analysis. To investigate the impact of mutation status on survival, we excluded 41 patients who had known residual disease at the completion of operation to include only those who underwent curative-intent R0 resections. Of the remaining 170 patients, 5 others were excluded because they died of other causes (n = 4) or of an unknown cause (n = 1)(Fig. 1).
Figure 1.
This flow chart outlines the selection and exclusion populations for the studied cohort.
Demographics and Tumor Characteristics
The mean age of the remaining 165 patients was 57.3 years (confidence interval [CI], 55.3–59.2 years), with a slight predilection toward male gender (55%). This cohort consisted of a heavily pretreated group, of which 72% had received preoperative chemotherapy and 20% had undergone previous liver resection. Key pathologic features included a mean tumor size of 4.3 cm (CI, 3.8–4.7 cm) and a disease-free interval <1 year (Table 1).
TABLE 1.
Patient Characteristics and Pathologic Variables, n = 165
| Characteristic | Value |
|---|---|
| Age: Mean [CI], y | 57.3 [55.3–59.2] |
| Men: No. (%) | 91 (55) |
| Synchronous disease: No. (%) | 30 (18) |
| Disease-free interval: Mean [CI], mo | 8.7 [6.2–11.1] |
| No. of tumors: Median [range] | 2 [1–12] |
| Tumor size: Mean [CI], cm | 4.3 [3.8–4.7] |
| Preoperative CEA: Mean [CI], ng/mL | 81.6 [33.4–129.8] |
| Preoperative chemotherapy: No. (%) | 91 (72) |
| Previous liver resection: No. (%) | 26 (19) |
Abbreviations: CEA, carcinoembryonic antigen; CI, confidence interval.
Mutation Status
Of all 165 patients, 50.6% had a single identified mutation, 30% had 2 or more mutations identified, and 19.4% had no identifiable mutation. The most common mutation was in TP53 (57.5%), followed by K/NRAS (43%; KRAS, n = 65; NRAS, n = 6). Other less frequent mutations included PIK3CA (12.1%) and BRAF (3%). The location of mutation in the K/NRAS gene was most often in exon 2 (codon 12 or 13, including G12D, G12S, G12V, G13C, G13D; 81.6% of RAS mutations), followed by exon 3 (codon 61, including Q61R and Q61H; 10% of RAS mutations), and exon 4 (codon 146, including A146T; 8.5% of RAS mutations) (Table 2).
TABLE 2.
Mutation Data, N = 165
| Mutation Site | No. of Patients(%) |
|---|---|
| No detected mutation | 32 (19.4) |
| KRAS/NRAS | 71 (43) |
| Exon 2 | 58 (35.1) |
| Exon 3 | 7 (4.2) |
| Exon 4 | 6 (3.6) |
| PIK3CA | 20 (12.1) |
| BRAF | 5 (3) |
| TP53 | 95 (57.5) |
Abbreviations: BRAF, v-Raf murine sarcoma viral oncogene homolog B1; KRAS, Kirstin rat sarcoma viral oncogene homolog; NRAS, neuroblastoma rat sarcoma viral oncogene homolog; PIK3CA, phosphoinositide 3-kinase catalytic subunit α; TP53, tumor protein 53.
Pathologic and Demographic Details by Mutation Status
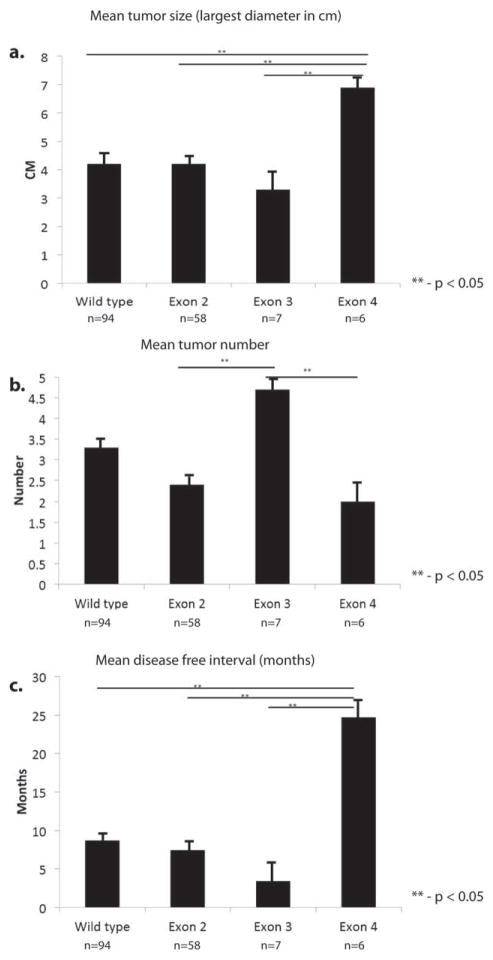
Demographic details, such as age and sex, did not differ significantly between patients who had different mutations or mutation locations within the gene. There were also no differences in mutation patterns between patients who did and did not receive preoperative chemotherapy. There were no statistically significant differences in pathologic features of the metastases (number or size) or lymph node status of the primary tumor by mutated gene (ie, wild type vs K/NRAS vs PIK3CA, etc); however, location of the K/NRAS mutation within the gene was associated with various pathologic tumor features. Patients who had tumors with exon 2 mutations (n = 58) had features similar to patients without K/NRAS mutation, with a mean size of 4.17 cm and an average of 2.4 tumors per resection. Those who had with mutations in exon 3 (n = 7) had a significantly greater number of tumors (mean, 4.7 tumors), which tended to be smaller (mean, 3.34 cm) and occurred at an earlier disease-free interval (measured from the date of liver resection) after resection of the primary tumor (mean, 3.4 months). Patients who had mutations in exon 4 (n = 6) tended to have larger (mean, 6.7 cm) solitary tumors that occurred after a longer disease-free interval (mean, 24.7 months) (Fig. 2).
Figure 2.
Kirstin rat sarcoma viral oncogene homolog/neuroblastoma rat sarcoma viral oncogene homolog (K/NRAS) mutation location is associated with varied tumor phenotype. Patients with exon 4 mutations had (a) larger tumors and (c) a longer disease free interval compared with those who had mutations located elsewhere. (b) Exon 3 mutation was associated with greater number of tumors.
Overall and Recurrence-Free Survival by Mutation Status
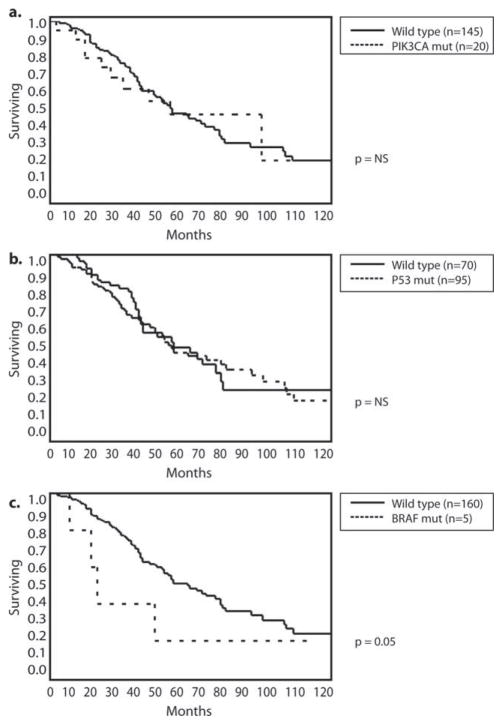
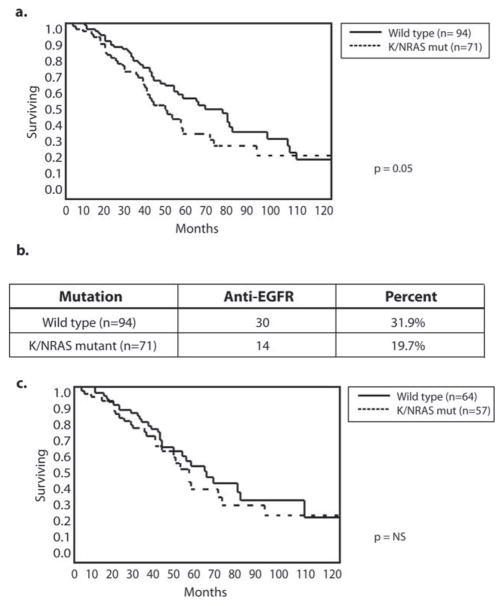
At a median follow-up of 45 months, 49% of patients had died of disease, 20% remained alive with active disease, and 31% remained alive with no evidence of disease. The 3-year and 5-year disease-specific survival (DSS) rates for the whole cohort were 69% and 48%, respectively, and the 3-year and 5-year recurrence-free survival rates were 30% and 26%, respectively. There were no differences in 5-year DSS between patients who had TP53 and PIK3CA mutations (5-year DSS, 47% and 50%, respectively) and those who had no mutations (Fig. 3a,b). Patients who had BRAF mutations had a significantly shorter 5-year DSS at 20% (Fig. 3c). Patients who had K/NRAS mutations had worse 3-year and 5-year DSS compared with those who had no mutations (60% vs 76% and 38% vs 54%, respectively; P <.05) (Fig. 4a). To account for the potential confounding effect of anti-EGFR therapies, next, we excluded patients who had received anti-EGFR treatment in either the preoperative or postoperative periods. Among the whole cohort of 165 patients, 44 had received either cetuximab or panitumumab, including 14 patients with and 30 patients without K/NRAS mutations. When those patients were excluded from the analysis, the 3-year and 5-year DSS rates of patients who had K/NRAS mutations were no different from the rates of patients without mutations (Fig. 4c).
Figure 3.
Kaplan-Meier curves illustrate overall survival for patients with or without (a) phosphoinositide 3-kinase catalytic subunit α (PIK3CA), (b) tumor protein 53 (P53), and (c) v-Raf murine sarcoma viral oncogene homolog B1 (BRAF) mutation (mut) status.
Figure 4.
Overall survival is illustrated according to Kirstin rat sarcoma viral oncogene homolog/neuroblastoma rat sarcoma viral oncogene homolog (K/NRAS) mutation status. (a) Kaplan-Meier curves indicate a difference in survival according to K/NRAS status. (b) Receipt of antiepidermal growth factor receptor (anti-EGFR) therapy is illustrated according to mutation status. (c) After the exclusion of those who received anti-EGFR therapy, there was no longer a difference in survival according to K/NRAS status.
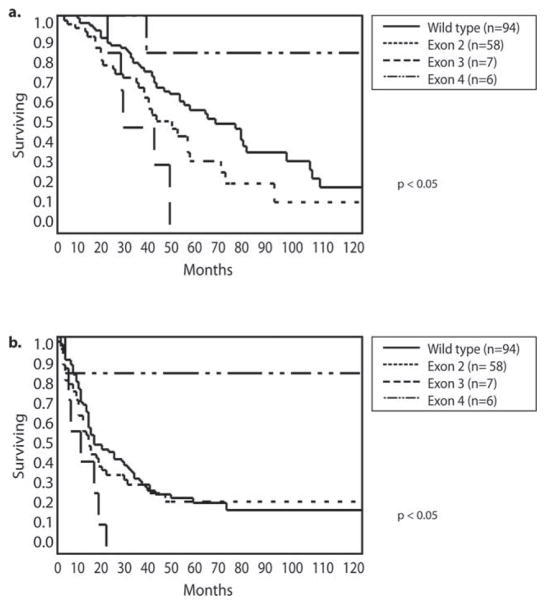
Next, we investigated the prognostic impact of different locations of K/NRAS mutations within the gene. The 6 patients who had mutations in exon 4 had a significantly better 5-year DSS (83%) than those who had mutations in exon 2 (35%) and those who had no K/NRAS mutation (54%; P <.05). There was only 1 death among these 6 patients at 35 months; the other 5 patients were alive at 47, 72, 165, 183, and 229 months. The 7 patients who had mutations in exon 3 had the worst outcomes, with no patients surviving at 5 years (Fig. 5a). The exclusion of patients who received anti-EGFR therapy had no impact on these results. A similar pattern was observed for recurrence-free survival: only 1 patient with an exon 4 mutation had a postoperative recurrence compared with all of those who had exon 3 mutations. The median time to recurrence for patients with exon 3 mutations was 12 months, whereas those who had either no mutation or the more common exon 2 mutations recurred at a median of time of 20 and 18 months, respectively (Fig. 5b).
Figure 5.
(a) Overall survival and (b) recurrence-free survival are illustrated according to Kirstin rat sarcoma viral oncogene homolog/neuroblastoma rat sarcoma viral oncogene homolog (K/NRAS) mutation location.
To exclude potential confounding, multivariate analysis was performed in which we included the variables associated with recurrence-free survival on univariate analysis as well as those previously identified as potential confounders (tumor size and number, disease-free interval, lymph node status of the primary tumor, previous chemotherapy, and preoperative carcinoembryonic antigen level). Multivariate analysis identified only size of the metastasis and driver mutation as predictors of 5-year recurrence-free survival with a risk ratio of 7.73 (CI, 1.3–154; P = .02) and 0.33 (CI, 0.15–0.88;P = .03) for RAS exon 4 and exon 3 mutations, respectively, compared with no RAS mutation.
Patterns of Recurrence
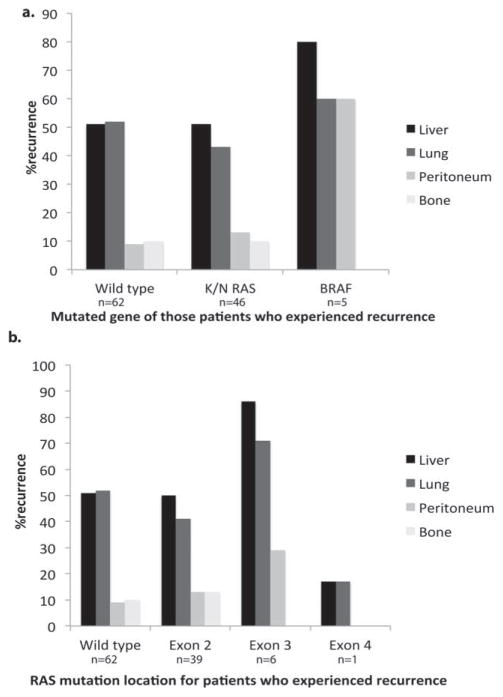
The gene and location of mutations were associated with the pattern of recurrence. Patients who had BRAF mutations recurred in a multifocal pattern, which included the lung, liver, and peritoneum. Those who had K/NRAS mutations recurred in the lung and liver with frequencies similar to those observed in patients who had wild-type K/NRAS. This contradicts previous reports suggesting that patients who have K/NRAS mutations more frequently recur with pulmonary metastases.14 When comparing recurrence patterns by location of K/NRAS mutation, patients who had exon 2 mutations relapsed in a pattern similar to that observed in those who had wild-type K/NRAS, with an equal proportion of liver and lung lesions. Patients with exon 3 mutations had a more diffuse recurrence pattern that included liver, lung, and peritoneal disease (Fig. 6).
Figure 6.
The incidence of recurrence location is illustrated according to (a) the mutated gene and (b) location of mutation on the K/NRAS gene in patients who developed recurrent disease. BRAF indicates v-Raf murine sarcoma viral oncogene homolog B1.
DISCUSSION
Colorectal cancer remains a significant cause of cancer-related mortality in the United States and worldwide. Patients often present with or develop metastatic disease, most commonly in the liver or lung. Metastasectomy for stage IV colon cancer has become an important tool in the treatment of patients who have limited metastatic disease to the liver or lung and is associated with prolonged survival and, occasionally, long-term cure.2,15,16 Despite advancements in preoperative prognostic modeling, we are still unable to predict clinically relevant differences in outcome that can dictate therapeutic decision making.6 However, as our insight into the biology of colorectal cancer improves, so too should our ability to prognosticate.
One of the most important advancements in our understanding of colon cancer biology was the discovery that a large proportion of tumors were driven by mutations located in the MAPK pathway, most notably KRAS.7 Since its discovery, the most clinically relevant finding related to RAS-mutated tumors has been its ability to determine eligibility for treatment with anti-EGFR–based therapy. More recently, RAS mutations have been studied for their potential to prognosticate recurrence and survival, providing a link between genetic mutation and biologic behavior.
Etienne-Grimaldi et al17 identified 93 patients with stage IV colon cancer who received fluorouracil-based chemotherapy and sequenced exon 2 of the KRAS gene. Those investigators identified mutations in 38.7% of patients and observed no correlation between RAS mutation status and response or survival. Contrary to this, Span et al18 demonstrated that survival in patients with all stages of colorectal cancer was significantly longer for those who had wild-type KRAS compared with KRAS mutations. Some of these discordant findings may relate to the way in which mutations in the KRAS gene are identified. Because a majority of mutations are identified in exon 2, many researchers and clinical centers only sequence this portion of the gene. Recent data have indicated that mutations in exons 3 and 4 as well as those in the related NRAS gene occur in an additional 10% of patients.13 Consequently, some patients who have RAS mutations are inadvertently grouped with those who do not have RAS mutations. Little is known about the biologic significance of various mutation locations within the RAS gene, although in vitro data suggest variable potential for activating downstream signaling, which may result in differing biology.13 Data linking survival and phenotypic characteristics of tumors to more in-depth mutational analysis are lacking.
To study this, we chose a population of patients who had undergone complete resection of CRLM. These patients, as a group, have heterogeneous outcomes, with both a high risk of recurrence and the potential for a cure. The inclusion only of those patients who underwent curative-intent R0 resection removes the confounder of disease burden to gain a clearer view of how driver mutations affect the biology of disease. A cohort similar to this has been the subject of 2 recent studies. Vauthey et al14 used a similar method of mutation capture and investigated the impact of KRAS mutation on survival and recurrence after hepatic metastasectomy. Those authors reported significantly worse survival among patients who had KRAS mutations compared with nonmutated controls. One key limitation to their study was that the incidence of KRAS mutation was only 18%, which is far lower than that reported by other groups. A second study by Karagkounis et al19 reported similar results, with worse overall and recurrence-free survival among patients who had KRAS mutations compared with those who were mutation-free. That study also excluded patients who received preoperative anti-EGFR therapy, and analyzed only exon 2 mutations, and had no capture of NRAS or exon 3 and 4 mutations. A recent report by Kemeny and colleagues examined both survival and recurrence patterns after hepatectomy according to KRAS mutation status and observed overall worse survival and a distinct pattern of recurrence. Like previous findings, nonexon 2 mutations were not routinely captured, and the study included only patients who had received hepatic arterial-directed chemotherapy, limiting the interpretation of results.20
In the current study, an analysis of MAPK pathway mutations in 165 patients who underwent curative-intent R0 resection of CRLM was undertaken to determine the prevalence and clinical significance of individual mutations. Sequenom mass spectrometry analysis was used to identify point mutations in exons 2, 3, and 4 of KRAS and in exons 2 and 3 of NRAS, BRAF and PIK3CA. Clinical, pathologic, and mutation data were available for 165 patients with a median follow-up of 45 months. There were no survival or pathologic differences in patients who had PIK3CA or TP53 mutations.
Patients with BRAF mutations were uncommon in this cohort and fared poorly, as previously reported.21,22 They tended to recur early and diffusely, and none were disease free at 2 years (data not shown). The incidence of mutation was only 3%, which was significantly lower than that reported by studies of primary tumors21 but in line with reports of metastases,12,14,19,23 highlighting the rarity with which these patients present with resectable disease.
The incidence of K/NRAS mutation in our cohort was 43%, which was higher than prior studies of hepatic metastasectomy,14,19 but in line with other reports of metastatic lesions.17,18 In our more in-depth investigation of the K/NRAS gene, approximately 20% of these mutations occurred in less common locations on exons 3 and 4. This is important, because these mutations are often excluded from traditional mutational analysis. When analyzing the whole cohort, K/NRAS mutation was associated with more frequent recurrence and worse DSS,20 as we previously reported. However, after eliminating patients who had received anti-EGFR therapy before resection or at the time of recurrence, this difference no longer existed, suggesting that the survival difference may reflect the efficacy of this therapy for K/NRAS nonmutants.
On the basis of in vitro data, we hypothesized that the location of a mutation within the K/NRAS gene may have an impact on tumor biology, recurrence, and survival. Janakiraman et al13 analyzed cell lines that had KRAS mutations in either exon 2, 3, or 4. Those authors observed that mutations in exons 2 and 3 resulted in more robust downstream signaling and, theoretically, more malignant potential compared with exon 4 mutations. To study this in vivo, we grouped patients according to the site of their K/NRAS mutation. In concert with the in vitro findings, patients who had mutations in exon 4 had significantly fewer recurrences and an improved DSS compared with those who had other sites of mutation. It is noteworthy that the patients who had exon 3 mutations fared poorly, all recurring within 2 years, and none survived past 4 years. Multivariate regression confirmed these findings, although the small sample size limited the analysis, as evidenced by the large confidence intervals. Pathologic features also differed according to mutation location. Tumors with exon 4 mutations tended to be large and solitary, whereas those with exon 3 mutations were smaller and more numerous.
As genetic sequencing becomes faster, more reliable, and affordable, we are able to glean increasing amounts of information from resected tumors that can improve our ability to prognosticate and treat. This study reveals an even greater complexity than previously recognized, in which not only does the identity of the driver mutation impact the biology of disease but so too does the location of that mutation within the gene. Although validation of these findings is needed before clinical applicability, it is reasonable to predict that, in the near future, mutation status will be used in the same way as tumor size and multiplicity in guiding our decision to pursue increasingly aggressive metastasectomies.
Limitations of this study include its retrospective nature and the single-institution sample. We also focused only on hotspot mutations, which fails to take into account the many other mutations present in colon cancer. It is possible that unrecognized mutations might be acting in synergy with KRAS mutations to amplify the findings we observed. Some patients and tumors were excluded because of poor quality of the slides or excessive tumor necrosis. Finally, because many of these mutations occur infrequently, in-depth analyses were limited by the low number of patients in each group, particularly for patients with exon 3 and 4 K/NRAS mutated tumors.
In conclusion, we have demonstrated an association between tumor phenotype and specific gene mutations as well as the location of a mutation within the gene. This serves as early data to suggest a possible new tool to help prognosticate outcomes after metastasectomy and to aid in preoperative patient selection. Further study, including prospective collection of mutation status and analysis of larger populations, is needed.
Acknowledgments
FUNDING SUPPORT
No specific funding was disclosed.
Footnotes
CONFLICT OF INTEREST DISCLOSURES
Nancy E. Kemeny reports grants from Amgen outside the submitted work. The remaining authors made no disclosures.
AUTHOR CONTRIBUTIONS
Timothy L. Frankel: Study concept and design, acquisition of data, analysis and interpretation of data, drafting of the article. Efsevia Vakiani: Study concept and design, acquisition of data, analysis and interpretation of data, and critical revision of the article for important intellectual content. Hari Nathan: Analysis and interpretation of data and critical revision of the article for important intellectual content. Ronald P. DeMatteo: Analysis and interpretation of data and critical revision of the article for important intellectual content. T. Peter Kingham: Analysis and interpretation of data and critical revision of the article for important intellectual content. Peter J. Allen: Analysis and interpretation of data and critical revision of the article for important intellectual content. Willliam R. Jarnagin: Analysis and interpretation of data and critical revision of the article for important intellectual content. Nancy E. Kemeny: Analysis and interpretation of data and critical revision of the article for important intellectual content. David B. Solit: Study concept and design, acquisition of data, analysis and interpretation of data, critical revision of the article for important intellectual content, and obtained funding. Michael I. D’Angelica: Study concept and design, acquisition of data, analysis and interpretation of data, critical revision of the article for important intellectual content, obtained funding, and study supervision.
References
- 1.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 2.Tomlinson JS, Jarnagin WR, DeMatteo RP, et al. Actual 10-year survival after resection of colorectal liver metastases defines cure. J Clin Oncol. 2007;25:4575–4580. doi: 10.1200/JCO.2007.11.0833. [DOI] [PubMed] [Google Scholar]
- 3.Nordlinger B, Sorbye H, Glimelius B, et al. Perioperative FOL-FOX4 chemotherapy and surgery versus surgery alone for resectable liver metastases from colorectal cancer (EORTC 40983): long-term results of a randomised, controlled, phase 3 trial. Lancet Oncol. 2013;14:1208–1215. doi: 10.1016/S1470-2045(13)70447-9. [DOI] [PubMed] [Google Scholar]
- 4.Gallagher DJ, Kemeny NE. Colorectal hepatic metastases: adjuvant chemotherapy and survival. J Clin Oncol. 2009;27:e18–e19. doi: 10.1200/JCO.2009.22.4626. author reply e20–e21. [DOI] [PubMed] [Google Scholar]
- 5.Kemeny NE. Treatment of metastatic colon cancer: “the times they are A-changing”. J Clin Oncol. 2013;31:1913–1916. doi: 10.1200/JCO.2013.49.4500. [DOI] [PubMed] [Google Scholar]
- 6.Zakaria S, Donohue JH, Que FG, et al. Hepatic resection for colorectal metastases: value for risk scoring systems? Ann Surg. 2007;246:183–191. doi: 10.1097/SLA.0b013e3180603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 8.Wan PT, Garnett MJ, Roe SM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 9.Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers [serial online] Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 10.Bokemeyer C, Bondarenko I, Makhson A, et al. Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the first-line treatment of metastatic colorectal cancer. J Clin Oncol. 2009;27:663–671. doi: 10.1200/JCO.2008.20.8397. [DOI] [PubMed] [Google Scholar]
- 11.Van Cutsem E, Kohne CH, Hitre E, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408–1417. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 12.Vakiani E, Janakiraman M, Shen R, Sinha R, Zeng Z, Shia J, et al. Comparative genomic analysis of primary versus metastatic colorectal carcinomas. J Clin Oncol. 2012;30:2956–2962. doi: 10.1200/JCO.2011.38.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janakiraman M, Vakiani E, Zeng Z, et al. Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Res. 2010;70:5901–5911. doi: 10.1158/0008-5472.CAN-10-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vauthey JN, Zimmitti G, Kopetz SE, et al. RAS mutation status predicts survival and patterns of recurrence in patients undergoing hepatectomy for colorectal liver metastases. Ann Surg. 2013;258:619–626. doi: 10.1097/SLA.0b013e3182a5025a. discussion 626–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fong Y, Fortner J, Sun RL, Brennan MF, Blumgart LH. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: analysis of 1001 consecutive cases. Ann Surg. 1999;230:309–318. doi: 10.1097/00000658-199909000-00004. discussion 318–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nordlinger B, Guiguet M, Vaillant JC, et al. Surgical resection of colorectal carcinoma metastases to the liver. A prognostic scoring system to improve case selection, based on 1568 patients. Association Francaise de Chirurgie. Cancer. 1996;77:1254–1262. [PubMed] [Google Scholar]
- 17.Etienne-Grimaldi MC, Formento JL, Francoual M, et al. K-Ras mutations and treatment outcome in colorectal cancer patients receiving exclusive fluoropyrimidine therapy. Clin Cancer Res. 2008;14:4830–4835. doi: 10.1158/1078-0432.CCR-07-4906. [DOI] [PubMed] [Google Scholar]
- 18.Span M, Moerkerk PT, De Goeij AF, Arends JW. A detailed analysis of K-ras point mutations in relation to tumor progression and survival in colorectal cancer patients. Int J Cancer. 1996;69:241–245. doi: 10.1002/(SICI)1097-0215(19960621)69:3<241::AID-IJC15>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 19.Karagkounis G, Torbenson MS, Daniel HD, et al. Incidence and prognostic impact of KRAS and BRAF mutation in patients undergoing liver surgery for colorectal metastases. Cancer. 2013;119:4137–4144. doi: 10.1002/cncr.28347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kemeny NE, Chou JF, Capanu M, et al. KRAS mutation influences recurrence patterns in patients undergoing hepatic resection of colorectal metastases. Cancer. 2014;120:3965–3971. doi: 10.1002/cncr.28954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roth AD, Tejpar S, Delorenzi M, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 2010;28:466–474. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 22.Farina-Sarasqueta A, van Lijnschoten G, Moerland E, et al. The BRAF V600E mutation is an independent prognostic factor for survival in stage II and stage III colon cancer patients. Ann Oncol. 2010;21:2396–2402. doi: 10.1093/annonc/mdq258. [DOI] [PubMed] [Google Scholar]
- 23.Santini D, Spoto C, Loupakis F, et al. High concordance of BRAF status between primary colorectal tumours and related metastatic sites: implications for clinical practice [serial online] Ann Oncol. 2010;21:1565. doi: 10.1093/annonc/mdq318. [DOI] [PubMed] [Google Scholar]