Table 3.
Experimental and computed binding free energies with and without salt effects for methyl octa-acid hosts and thermodynamic decompositions of binding free energies calculated with inclusion of salt effects
| Guests | Structure |
|
Without salt effects | With salt effects
|
|||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
ΔEbd |
|
ΔEreorgf |
|
||||
| G1 |
|
−5.3 | −7.5 ± 0.06 | −4.8 ± 0.08 | −17.2 ± 0.01 | 12.4 ± 0.08 | 0.8 ± 1.4 | 11.5 ± 1.4 | |
| G2 |
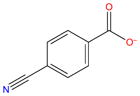
|
−5.1 | −10.6 ± 0.07 | −7.7 ± 0.09 | −17.6 ± 0.01 | 9.9 ± 0.1 | 1.4 ± 1.7 | 8.5 ± 1.6 | |
| G3 |
|
−6 | −16.2 ± 0.07 | −9.4 ± 0.1 | −21.4 ± 0.01 | 12 ± 0.1 | 0.7 ± 1.7 | 11.3 ± 1.7 | |
| G4 |
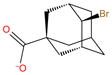
|
−2.4 | −2.4 ± 0.1 | −0.7 ± 0.2 | −21.1 ± 0.02 | 20.4 ± 0.2 | 6.7 ± 2.7 | 13.7 ± 2.7 | |
| G5 |
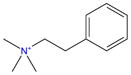
|
−3.9 | −14.5 ± 0.07 | −5.7 ± 0.1 | −16.5 ± 0.02 | 10.8 ± 0.1 | 1.2 ± 1.8 | 9.6 ± 1.8 | |
| G6 |
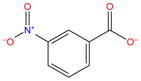
|
−4.5 | −8.9 ± 0.06 | −5.8 ± 0.09 | −18.0 ± 0.02 | 12.2 ± 0.1 | 1.4 ± 1.6 | 10.8 ± 1.6 | |
| RMSE | 6.7 | 2.1 | |||||||
| Correlation coefficient (r) | 0.66 | 0.89 | |||||||
All values in kcal/mol.
Experimental ITC binding free energy, except for G4 and G5 for which only NMR measurements are available [35].
Predicted binding free energy without addition of ionic effects.
Binding free energy prediction with the addition of salt effects.
Average interaction energy of the bound complex at =1. The uncertainty is estimated as the standard error of the mean.
Reorganization free energy as calculated using Eq. 9,
Reorganization energy or intra-molecular strain as calculated using Eq. 10,
Binding entropy computed by finite difference method and evaluated at 300K; the uncertainty is computed by error propagation for all the decompositions
