Figure 2.
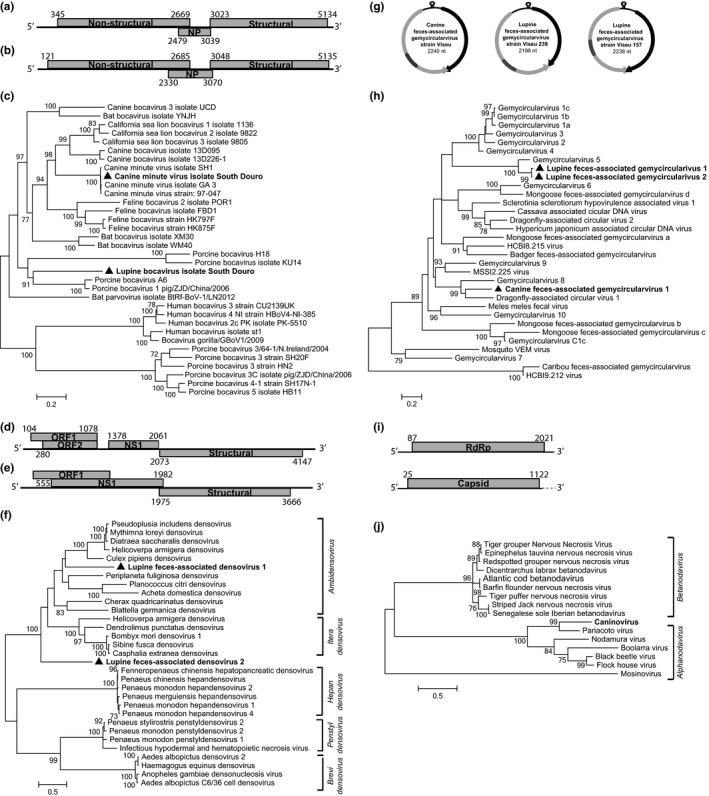
(a) Genome organization of the canine minute virus. (b) Genome organization of the lupine bocavirus. (c) Maximum‐likelihood phylogenetic tree of the aa VP1 sequences of bocaviruses. (d) Genome organization of the lupine feces‐associated densovirus 1. (e) Genome organization of the Lupine feces‐associated densovirus 2. (f) Maximum‐likelihood phylogenetic tree of the aa NS1 sequences of densoviruses. (g) Genome organization of undescribed gemycircularviruses. Depicted in gray is the Rep with an intron and depicted in black is the capsid protein. (h) Maximum‐likelihood phylogenetic tree of the aa replicase sequences of gemycircularviruses. (i) Genome organization of the caninovirus showing a complete RNA1 segment and a partial RNA2. (j) Maximum‐likelihood phylogenetic tree of the aa RNA2 (Capsid) gene sequences of nodaviruses. All trees were built in MEGA6 using 500 bootstraps, and only bootstrap values even or >70 are shown. Strain identified in this study is shown in bold with a triangle
