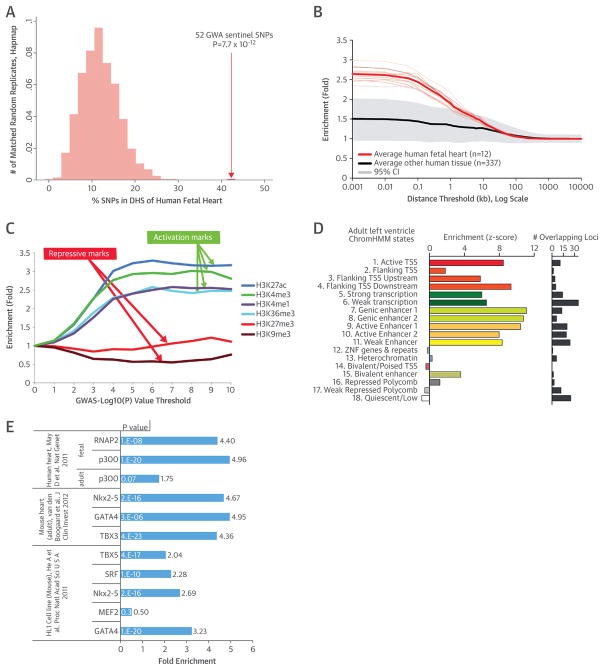
FIGURE 2. Functional Annotations.
(A) The 52 sentinel SNPs are significantly enriched in deoxyribonuclease hypersensitivity sites (DHS) of the human fetal heart compared with the matched random distribution of HapMap SNPs. (B) The effect of physical distance between SNPs that meet genome-wide significance (p < 1 × 10−8) on enrichment of fetal heart relative to all other tissues at DHS. The enrichment is strongest at the SNP’s location and decreases after 100 base pairs from the SNP sites. (C) SNPs associated with QRS traits are enriched for the activating histone modifications H3K27ac, H3K4me3, H3K4me1, and H3K36me3 in the human left ventricle, which increased at more stringent genome-wide association study (GWAS) p value thresholds. The repressive mark H3K27me3 is not enriched, whereas H3K9me3 is significantly reduced, suggesting that QRS-trait loci are predominantly expressed in the left ventricle. (D) To capture greater complexity, we performed an integrative analysis in an 18-state “expanded” ChromHMM model representative of different functional regions of the genome. The 52 loci for the 18-state model were enriched using the 6 core histone marks (left); the total number of the 52 loci overlapped by each feature is shown (right). (E) SNPs (p < 1 × 10−8) were also significantly enriched for various factors in the human heart, mouse heart, and the HL-1 cell line. CI = confidence interval; TSS = transcription start site; other abbreviations as in Figure 1.