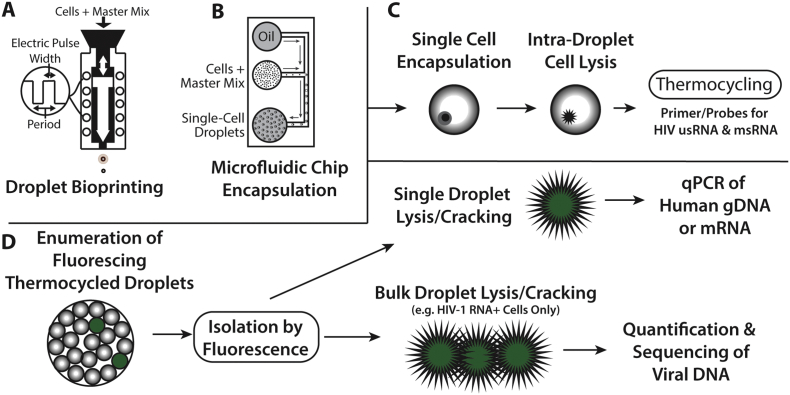
Fig. 1.
Schema of the single cell encapsulation, lysis, HIV-1 detection, and rescue of cellular genomic DNA and mRNA. Cells are encapsulated in a master mix including PCR enzymes, primers, probes, and cell lysis agents (Supplementary Table 1) into oil by either brioprinting or microfluidic chip encapsulation (A, B) as described in the methods. Up to 20,000 droplets are then added to each well of a 96 well PCR plate. Cells are lysed within isolated droplet microenvironenments followed by PCR amplification of Tat-Rev spliced or unspliced cell-associated HIV-1 RNA (C). Cells are contained in the hydrophilic droplets which are stabilized by the oil-droplet interface. Droplets containing infected cells can be enumerated using an oil-based commercial flow cytometer or by direct visualization followed by microfluidic sorting of positive droplets (D). HIV-1 RNA positive or negative cells can be isolated using ultrafine needle aspiration, placed into individual microwell tubes or plate wells followed by droplet “cracking” (i.e. lysing droplets to release nucleic acids) and further characterized. In this study, human or viral genomic DNA and human mRNA from droplets containing a single encapsulated cell or from bulk droplets containing HIV-1 RNA positive cells were quantified or sequenced.