Abstract
Precisely targeted genome editing is highly desired for clinical applications. However, the widely used homology-directed repair (HDR)-based genome editing strategies remain inefficient for certain in vivo applications. We here demonstrate a microhomology-mediated end-joining (MMEJ)-based strategy for precisely targeted gene integration in transfected neurons and hepatocytes in vivo with efficiencies up to 20%, much higher (up to 10 fold) than HDR-based strategy in adult mouse tissues. As a proof of concept of its therapeutic potential, we demonstrate the efficacy of MMEJ-based strategy in correction of Fah mutation and rescue of Fah−/− liver failure mice, offering an efficient approach for precisely targeted gene therapies.
Keywords: MMEJ, CRISPR/Cas9, In vivo targeted integration, Fah−/− mice, Gene therapy
Highlights
-
•
MMEJ-based strategy showed much higher knock-in (up to 10 fold) efficiencies than HDR-based strategy in adult mouse tissues.
-
•
MMEJ-based strategy proved the efficacy in correction of Fah mutation and rescue of Fah−/− liver failure mice.
1. Introduction
The CRISPR/Cas9 technology provides a promising tool for genetic engineering, opening up the possibility of correcting genetically mutated cells and tissues in situ (Komor et al., 2017, Yin et al., 2014, Yin et al., 2016). Targeted genome editing has generally been achieved by homology-directed repair (HDR)-based genome editing strategies, allowing precise insertion of large DNA fragments. However, in vivo precisely targeted integration is still infeasible due to low HDR efficiency. Alternatively, non-homologous end joining (NHEJ), another major double strand break (DSB) repair pathway, has been demonstrated facilitating gene knock-ins in cultured cells, embryos, and more importantly in vivo (Maresca et al., 2013, Kimura et al., 2014, Suzuki et al., 2016). Using this method with programmable nucleases, a targeted genomic and a donor vector with artificial recognition sequences are simultaneously cleaved and ligated to each other, presumably through NHEJ. However, NHEJ repair system introduces various types of indel mutations at the junctions and thus these targeted integrations are ‘simple ligations’ (Kimura et al., 2014, Auer et al., 2014, Nakade et al., 2014, Hisano et al., 2015, Li et al., 2015, Cristea et al., 2013, Maresca et al., 2013).
By contrast, the other method for achieving precise integration was reported via microhomology-mediated end-joining (MMEJ)-based strategy. In this method, a donor DNA contained gRNA target sequences as well as microhomologous sequences (5–25 bp) for error-prone end-joining. MMEJ-dependent targeted integration has been demonstrated in culture cells and generation of gene-modified animals (Nakade et al., 2014, Hisano et al., 2015). Nevertheless, application of MMEJ in tissues in vivo has yet to be demonstrated. In this study, we demonstrated that MMEJ-mediated targeted integration was a robust knock-in strategy both ex vivo and in vivo, and could be used for correction of Fah mutation in hepatocytes, reducing liver damage and rescue Fah mutated mice from death.
2. Materials and Methods
2.1. Animals
All procedures were approved by the Animal Committee of the Institute of Neuroscience, Chinese Academy of Sciences. Mice were housed and bred in a 12-h light-dark cycle in the animal facility of the Institute of Neuroscience. C57BL/6 mice were purchased from Slac Laboratory Animals (Chinese Academy of Sciences). Fah−/− mice were kept on 10 mg/L NTBC water.
2.2. Construction of Plasmids
To clone a single Cas9-sgRNA-EGFP expressing vector, a modified pX330 (Addgene catalog no. 42230) expression vector expressing Cas9-CMV-EGFP and sgRNA was linearized with BbsI digestion, and gel purified. A pair of oligos for each targeting site were phosphorylated, annealed, and ligated to the linearized pX330.
To construct HDR donor for mouse Actb gene (see “Plasmid-based donor template sequences”), mCherry, EF1a-EGFP, 5′ and 3′ homology arms (800 bp) were amplified from pAAV-Ef1a-DIO-mCherry-WPRE-pA (Addgene catalog no.37083), CAG-GFP-IRES-CRE (Addgene catalog no.48201) or mouse genome, then subcloned donor (800bpHAL-p2A- mCherry-800bpHAR), U6-Actb-sgRNA expression cassette and EF1a-EGFP expression cassettes between ITRs of pAAV vector (Addgene catalog no.37083).
To construct MMEJ donor for Actb gene (see “Plasmid-based donor template sequences”), donor DNA (HAL-p2A-mCherry-HAR) sandwiched by 23 nt Actb-sgRNA target sequence, U6-Actb-sgRNA expression cassette and EF1a-EGFP expression cassettes were subcloned between ITRs of pAAV vector (Addgene catalog no.37083).
The resulting fragment, or linearized vector were purified with Gel Extraction Kit (Omega, D2500-02) and concentrated with ethanol precipitation. All the plasmid constructs were extracted with Plasmid Midi Kit (Qiagen, 12143) and verified with DNA sequencing.
2.3. Surveyor Assay
The different samples collected by FACS were digested in lysis buffer (0.1% Triton X-100, 0.1% Tween 20, and 100 μg/mL Proteinase K) and incubated for 30 min at 56 °C and heat inactivate proteinase K at 95 °C for 10 min. The sample was amplified by primers around the sgRNA targeting site (Table S3). 600 ng wild-type purified PCR product and 600 ng of the tested purified PCR product were denatured and re-annealed in NEB Buffer 2 (New England Biolabs), respectively. The PCR products were digested with T7EN1 (M0302L, New England Biolabs) for 2 h at 37 °C and separated by 2% agarose gel electrophoresis.
2.4. Production of Lentivirus and Infection of Primary Cells
Lentivirus was packaged by transfecting HEK293T cells using Polyethylenimine (PEI) at a final concentration of 50 μg/mL, the ratio of MMEJ donor/sgRNA and packaging vectors psPAX2 (Addgene 12260) and pMD2.G (Addgene 12259) is 4:3:2, respectively. Virus supernatant was collected at 2rd to 3th day post-transfection. The titer of lenti-MMEJ donor/sgRNA and lenti-Cas9 was 4.45 ∗ 109 and 2.11 ∗ 109, respectively. Astrocytes and neurons were infected with a mixture of lenti- MMEJ donor/sgRNA and lenti-spCas9 virus.
2.5. Cell Culture and Transfection
Mouse ESCs cells (129/Sv × C57BL/6 ES cell and E14 cell) were cultured with 2i medium, consists of Dulbecco's modified Eagle's Medium (DMEM) (Gibco,11965-092) plus 15% fetal bovine serum (FBS) (Gibco), 1000 U/mL mouse Lif, 2 mM glutamine (Sigma), 1 mM sodium pyruvate (Sigma), 0.1 mM β-mercaptoethanol (Sigma), 0.1 mM non-essential amino acids (Gibco), 1 μM PD0325901 and 3 μM CHIR99021. N2A cells were cultured with DMEM plus (Gibco) 10% FBS (Gibco). All cells were cultured at 37 °C with 5% CO2 incubation. Mouse ESCs cells were transfected using Lipofectamine 3000 Reagent (Invitrogen) according to the manufacturer's instructions. For each well of a 6-well plate, a total of 5 μg plasmids (Cas9: MMEJ donor = 1:1) was used. After 48 h, transfection-positive ES cells were sorted into 6-well plates using BD FACS Aria II for further culture and analysis.
Primary cultures of astrocytes were prepared as described previously (Liu et al., 2015). Primary astrocytes were obtained from the dorsal midbrain of P5–P7 mice and were cultured with a medium consisting of DMEM/F-12, 10% fetal bovine serum (Invitrogen), penicillin/streptomycin (Invitrogen), and supplemented with B27 (Invitrogen), 10 ng/mL epidermal growth factor (EGF), and 10 ng/mL fibroblast growth factor 2 (FGF2). Primary neurons were obtained from the cortex of E14.5 C57 mouse brains and plated at a density of 2 ∗ 105 cells/well onto glass coverslips coated with poly-d, l-Lysine and pre-incubated in medium containing 5% fetal bovine serum. After 1 h, culture medium was changed to serum-free Neurobasal medium with 2% B27 (Invitrogen), 1% Glutamax (Invitrogen) and 1% penicillin/streptomycin (Thermo Fisher Scientific). All cells were cultured at 37 °C with 5% CO2 incubation. Half volume of culture media was replaced every 3 days.
2.6. Hydrodynamic Injection and Hepatocyte Isolation
Vectors for hydrodynamic tail-vein injection were prepared using the EndoFree-Midi Kit (Qiagen). For hydrodynamic liver injection, plasmid DNA suspended in 2 mL saline was hyperdynamically injected into 8-week-old male/female mice (C57BL/6J or Fah−/−) via the tail vein in 5–7 s. The amount of injected DNA was 30 μg each for HDR donor + spCas9, 30 μg each for MMEJ donor + spCas9. An equal amount of HDR donor only or MMEJ donor only was used as a control for each experiment. C57 mice were sacrificed at days 5–9 post injection. Separated liver lobes were harvested for either genomic DNA extraction or fixed with 4% of paraformaldehyde. For hepatocyte isolation, primary mouse hepatocytes were isolated by standard two-step collagenase perfusion method (Huang et al., 2011). Hepatocytes were purified by 40% Percoll (Sigma) at low-speed centrifugation (1000 rpm, 10 min).
2.7. In Utero Electroporation
The experimental procedures for in utero electroporation have been described previously (Takahashi et al., 2008). E14.5 pregnant C57BL/6 mice were anaesthetized with Pentobarbital sodium (50 mg/kg, Sigma). The final concentration of each plasmid (EFs-spCas9-NLS-SV40polyA, the donor vector for HDR and the donor for MMEJ were 2 μg/μl, respectively. Plasmids were injected into the embryos' lateral ventricles with 0.005% fast green solution (Sigma). For electroporation, 5 pulses with a 50-ms duration separated by 950-ms were applied at 35 V using ECM 830 (BTX). The uterine horns then were placed back into the abdominal cavity and allowed to develop in utero for the indicated time.
2.8. Immunostaining and Immunohistochemistry
For immunostaining, mice were transcardially perfused with 0.9% saline followed by 4% paraformaldehyde using a peristaltic pump (Gilson) and fixed overnight at 4 °C. Then the tissue was dehydrated using 30% sucrose until sinking to the bottom of tube. Tissue sectioning was performed on a vibratome (VT 1200S, Leica Microsystems) at a thickness of 40 μm for brain, 10 μm for liver. The brains were sectioned coronally at 50 μm with a vibratome (VT 1200S, Leica Microsystems). Sections were rinsed three times in 0.1 M phosphate-buffered (PB) and incubated with primary antibodies diluted in diluent with 5%NGS overnight at 4 °C. The following day, sections were washed three times in PB and then incubated with secondary antibody (1:400, Life Technologies) for 2 h at room temperature on an orbital shaker. Finally, the sections were counterstained with DAPI for 20 min and mounted with SlowFade Diamond Antifade Mountant (Life) on glass slides.
For immunohistochemistry, mouse liver tissues were fixed with 4% paraformaldehyde and embedded in paraffin. 3–4 μm paraffin sections were deparaffinized and rehydrated before staining. The sections were incubated with primary antibody: rabbit anti-FAH (1:1000, BangLv-MN055) overnight at 4 °C. The following day, sections were washed three times in PB and then incubated with Anti-rabbit secondary antibody conjugated with biotin for 1 h at room temperature, and then incubated with ABC kit (Vector, PK-6100) for 30 min at room temperature. DAB (Vectorlabs, SK-4105) staining was applied on the HRP-conjugated antibody incubated sections. The sections were counterstained in hematoxylin (Beyotime Biotechnology, C0105) and covered in neutral balsam (HUSHI, 10004160).
2.9. Serum Biochemistry
Blood was collected using retro-orbital puncture. Aspartate aminotransferase (AST) and alanine aminotransferase (ALT) levels in serum were determined using diagnostic assay (ELISA) kits (Bioflin, ml0373361 & ml037453).
2.10. Single-cell Genotyping Analysis
For picking up and transferring single cells or clones, we used a glass capillary under a dissection microscope. Single mESCs clones, Fah+ clones or single cells were picked up based on fluorescence or immunohistochemistry, and transferred directly into PCR tubes containing 1.5 μl embryo lysis buffer (0.1% tween 20, 0.1% Triton X-100 and 4 μg/mL proteinase K). The samples were incubated for 30 min at 56 °C and heat inactivate proteinase K at 95 °C for 10 min. PCR amplification was performed using nest primer sets (Table S3). ExTaq was activated at 95 °C for 3 min, and PCR was performed for 34 cycles at 95 °C for 30 s, 62 °C for 30 s, and 72 °C for 1 min, with a final extension at 72 °C for 5 min. Secondary PCR was performed using 0.5 μl primary PCR product and nested inner primer. PCR was carried out in the same reaction mixture. PCR product was gel purified and sequenced.
2.11. Statistical Analysis
All statistical values were presented as mean ± SEM. Differences between datasets were judged to be significant at p < 0.05.
3. Results
3.1. MMEJ-mediated Precise Integration In Vitro and Ex Vivo
We first tested whether MMEJ could mediate targeted integration of transgenes in cultured cells. We used modified precise-integration-into-target-chromosome (PITCh) vector (Nakade et al., 2014) as a base MMEJ plasmid. The MMEJ construct contained an insertion fragment flanked by sgRNA target sequences (23 bp) and short homologous sequences (20–40 bp) on both sides adjoining the double strand breaks (DSBs) (Fig. 1a). This construct was designed to fuse a p2A-mCherry reporter with the last codon of the Actb gene and the knock-in efficiencies were presented as percentages of mCherry+ cells (Fig. 1a). We first designed three sgRNAs targeting the loci surrounding last codon of the Actb gene (Fig. S1a). By T7E1 assay in N2A cells, we chose Actb-sgRNA-2 with 22.45% cutting efficiency for further study (Fig. S1a–b). Seven days after transfection, we observed successful targeted integrations in mouse embryonic stem cells (mESCs), as indicated by mCherry expression (Fig. S1c). Genotyping results showed that 11/11 and 8/11 of the examined mCherry+ clones were precise in-frame integrations in 5′ junctions and 3′ junctions, respectively, indicating MMEJ had achieved precise integration in mESCs (Fig. S1d–e). MMEJ-targeted integration was also verified in other genome loci, including Oct4, Rosa26 and Hist1h3a (Figs. S1f–j and S2; Table S1), with 3%–12% efficiency. MMEJ-targeted integration could be also achieved by a double-cut donor containing two universe sgRNAs other than two sgRNAs the same as endogenous site (Figs. S2g and S3; Table S1). This strategy may increase the knock-in efficiency at the locus with a relatively low sgRNA cutting efficiency, as previous reported (Kimura et al., 2014). We found very little to no knock-in events when using donor DNA with no cutting site (Fig. S3; Table S1). We also found that the knock-in efficiency of this strategy was not improved by L755507 or Scr7, which had been reported to improve the efficiency of HDR (Fig. S3; Table S1) (Maruyama et al., 2015, Chu et al., 2015, Yu et al., 2015). Notably, L755507 treatment decreased the knock-in efficiency (Fig. S3; Table S1), indicating the different mechanism between HDR-based method and MMEJ-based method.
Fig. 1.
MMEJ-mediated targeted integration in N2a cells, astrocytes and neurons. (a) Schematic overview of MMEJ-mediated gene targeting strategy by lentivirus in vitro. N2a cells, primary astrocytes or neurons were infected with lentivirus-mediated Cas9 plus each donor vector. (b) Representative immunofluorescence images of cells infected with lentivirus 5 days later. +/−, with/without lentivirus expressing Cas9. Scale bar, 100 μm. Arrowheads mark cells with mCherry or GFP, indicating successful integration of donors into targeted sites. (c) Relative knock-in efficiencies indicated by percentage of mCherry+ cells among transfected cells GFP+. Results were obtained from cell slices (blue dots) or FACS analysis (green dots). The input data points were shown as dots. Dots represent repeat times for each locus. (d–f) Genotyping analysis of MMEJ-mediated knock-in at Actb locus in N2a cells (d), astrocytes (e) or neurons (f). PCR products of transfected cells were sequenced. Mecp2 as reference gene. (g) Single cell analysis of mCherry+ cells after 2A-mcherry knock-in into Actb locus by MMEJ. PCR products amplified from 5′ and 3′ junction sites were sequenced. The MMEJ intended knocked-in sequence is shown at the top. Upper: micro-homology arm; underline, linker; purple, p2A; red, mCherry. The PAM sequence is highlighted in blue.
Next we tested whether MMEJ-based method could be achieved by lentivirus delivery method, which is widely used for in vivo applications. We infected N2A cells, mouse primary astrocytes and neurons, with MMEJ donor/sgRNA and Cas9 lentivirus for targeted integration at Actb locus. By the fifth day post-transfection, we observed that 4.5 ± 0.5%, 4.9 ± 0.4%, and 3.4 ± 1.6% of transfected N2A cells, astrocytes and neurons showed mCherry expression, respectively (Figs. 1a–c and S4). Single-cell sequencing analysis of individual mCherry+ cells showed that all examined integration events were precise in-frame integration at 5′ and 3′ junctions, indicating that MMEJ-mediated integration could be applied in highly proliferative N2A cells, as well as slowly-dividing primary astrocytes and non-dividing primary neurons (Fig. 1d–g). Overall, exogenous DNA could be precisely integrated into the genome by MMEJ-based strategy in vitro.
3.2. MMEJ-mediated Precise Integration In Vivo
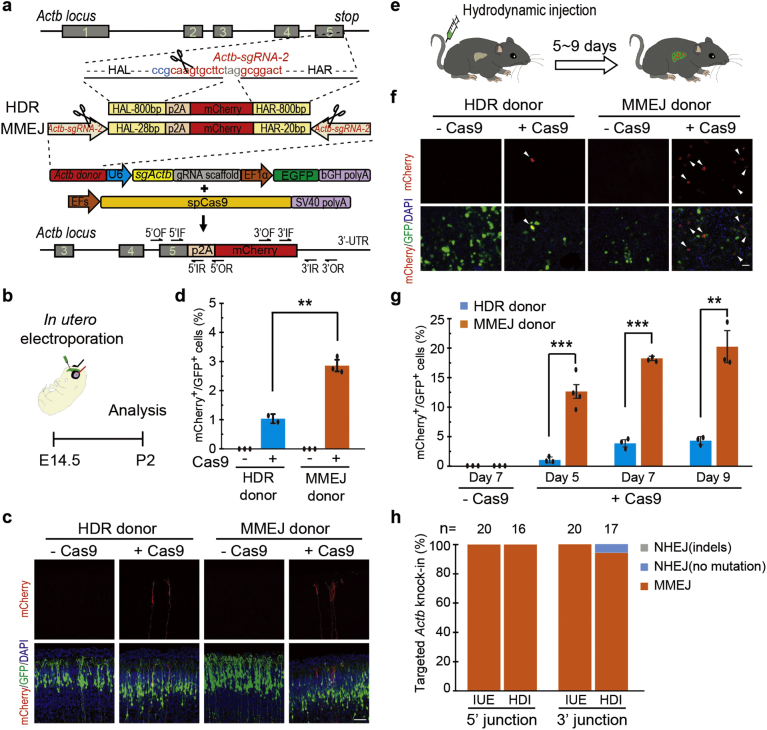
As confirmed by in vitro experiments, we set out to examine whether MMEJ-based method could be applied to in vivo DNA integration. We first delivered Actb-MMEJ constructs to E14.5 mouse brain via in utero electroporation (Fig. 2a–b). Seven days after electroporation, we observed that 3.61 ± 0.15% of electroporated cells (mCherry+/GFP+, relative efficiency) showed mCherry expression (Fig. 2c–d). As a comparison, only 0.83 ± 0.21% of electroporated cells with HDR donor were mCherry positive (Fig. 2a, c, d). We next delivered the MMEJ constructs to the mouse liver by tail-vein hydrodynamic injection (Fig. 2e). We observed that 12.85 ± 1.66%, 18.52 ± 0.37% and 20.52 ± 2.75% of transfected hepatocytes (mCherry+/GFP+, relative efficiency) showed mCherry expression at day 5, 7 and 9 post-injection, respectively (Fig. 2f–g). By contrast, only 1.16 ± 0.50%, 4.00 ± 0.63% and 4.45 ± 0.71% of transfected hepatocytes with HDR donor were mCherry+, respectively (Fig. 2f–g). MMEJ-based targeted integrations in mouse brain and hepatocytes were also confirmed by genotyping and DNA sequencing (Figs. 2h and S5a–d). We also counted knock-in efficiencies by the percentage of mCherry+ cells in total neurons or hepatocytes and observed the similar trends (Fig. S5e–f). Together, MMEJ-based method showed much higher DNA integration efficiency than HDR-based method in vivo.
Fig. 2.
In vivo genome editing via MMEJ-mediated targeted integration. (a) Schematic of HDR- and MMEJ-mediated gene targeting strategy for generating Actb-2A-mCherry knockin in vivo. The sgRNA target sequence is shown in red and the protospacer adjacent motif (PAM) sequence in blue. Black scissors, Cas9 cleavage site. Actb-sg is CRISPR/Cas9 target sites to create 28(20)-bp micro-homology arm at both ends of the p2A-mCherry cassette. HDR donor, genome cut-only. MMEJ donor, genome and donor cut-both. OF/OR, outer forward/reverse primer. IF/IR, inner forward/reverse primer. HAL, homology arm left. HAR, homology arm right. (b) Experimental scheme for targeted Actb-2A-mCherry knock-in in fetal brain via in utero electroporation. (c) Representative immunofluorescence images of neurons showing correct mCherry knock-in at the Actb locus. Scale bar, 50 μm. GFP, transfected cells. (d) Relative knock-in efficiency measured by the percentage of mCherry+ cells among GFP+ cells. Results were obtained from at least 2 mice and presented as mean ± s.d. The input data points were shown as black dots. **p < 0.01, unpaired Student's t-test. (e) Experimental scheme for targeted Actb-2A-mCherry knock-in via hydrodynamic tail vein injection. (f) Representative immunofluorescence images of hepatocytes in liver sections at Day 7 post injection. Scale bar, 50 μm. GFP, transfected cells. Arrowhead, mCherry+/GFP+ cell. (g) Relative knock-in efficiency measured by the percentage of mCherry+ cells among GFP+ cells. Hepatocytes were harvested at Day 5, 7 or 9 post injection. Results were obtained from at least 3 mice and presented as mean ± s.d. The input data points were shown as black dots. **p < 0.01, ***p < 0.001, unpaired Student's t-test. (h) The fidelity of 5′ and 3′ junction sites of single mCherry+ cells after 2A-mCherry knock-in at Actb locus by MMEJ. n, number of analyzed cells. MMEJ, precise integration; NHEJ (no mutation), integration with additional microhomology arm; NHEJ (indels), integration with random mutations. IUE, in utero electroporation; HDI, hydrodynamic injection.
3.3. MMEJ-mediated Gene Therapy in Fah−/− Mice
To explore the possibility of using MMEJ for gene replacement therapy, we used fumarylacetoacetate hydrolase (Fah)-deficient mice, a hereditary tyrosinemia type I (HTI) mouse model (Azuma et al., 2007, Paulk et al., 2010). HTI is caused by an inborn error of metabolism resulting from the mutation of Fah, the enzyme for the last step of tyrosine catabolism. Lack of Fah expression leads to accumulation of hepatotoxins in the hepatocytes, thus causing hepatic injury and liver failure. Fortunately, Fah−/− mice could be maintained by administration with an inhibitor of upstream of tyrosine catabolic pathway, 2-(2-nitro-4-trifluoromethylbenzoyl)-1,3-cyclohexanedione (NTBC) (Paulk et al., 2010). The Fah−/− mouse we used here harbored an insertion in exon 5 of Fah gene, which caused a frameshift mutation (Grompe et al., 1993) (Fig. 3a). Thus we set out to see if MMEJ-mediated gene correction could rescue Fah mutation in the Fah−/− mouse.
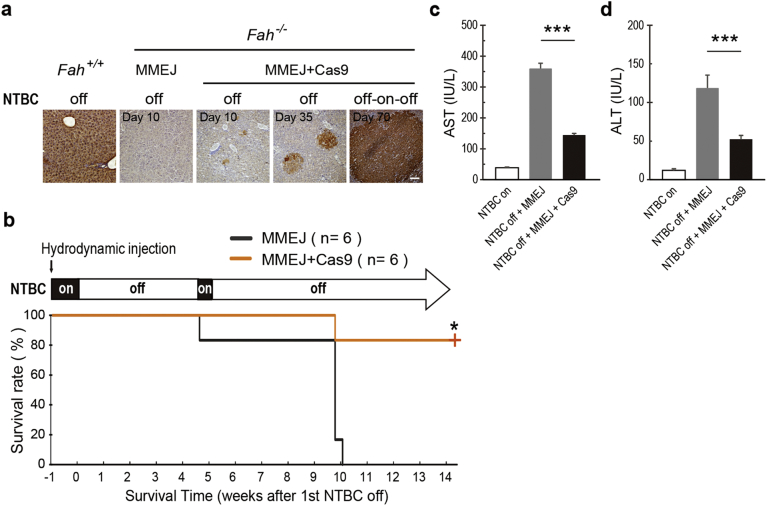
Fig. 3.
MMEJ-mediated gene correction of Fah-deficient mice. (a&b) Schematic representation of MMEJ gene replacement strategy on Fah-deficient mice. CRISPR/Cas9 components and Fah-MMEJ construct, designed to insert Fah cDNA of exon 5 to 14 into intron 4 of Fah gene, were delivered into Fah−/− mouse livers by hydrodynamic injection. NTBC on: Fah−/− mice were kept on NTBC water; NTBC off: withdrawal of NTBC water (defined as day 0, which is NTBC on 7 days after injection). (c) Fah immunostaining of Fah−/− mice injected with MMEJ donor or MMEJ donor plus Cas9. Scale bar, 50 μm. (d) The fidelity of 5′ and 3′ junction sites of single Fah+ clone after gene correction in Fah-deficient mice. n, number of analyzed clones. (e) Body weight of Fah−/− mice injected with MMEJ donor or MMEJ donor plus Cas9. Body weight was monitored over time and normalized to pre-injection weight. MMEJ only, n = 6; MMEJ + Cas9, n = 6.
We first designed three sgRNAs targeting the intron 4 of the Fah gene and Fah-sgRNA-1 with 37.79% cutting efficiency was selected based on T7E1 assay (Fig. S6a). We hydrodynamically injected Cas9 construct together with Fah-MMEJ constructs, designed to insert Fah cDNA of exon 5 to 14 into intron 4 of Fah gene, to Fah−/− mouse livers (Fig. 3a). One week after injection, NTBC was withdrawn to induce liver damage (Fig. 3b). Remarkably, hepatocytes expressing corrected Fah were found after one-week-withdrawal of NTBC (Figs. 3c and S6b–c). According to the sequencing results, we confirmed the successful correction of the Fah gene in hepatocytes (Figs. 3d and S6d–e). Note, in contrast to 5′ junctions with precise integration, the majority of targeted integrations with Fah-MMEJ constructs at 3′ junctions showed non-precise integration, possibly due to NHEJ-mediated repair rather than MMEJ-mediated repair. The difference between 5′ junctions and 3′ junctions may be caused by the detection method we used. We picked up the single knock-in positive clone (Fah+) to do with the DNA sequencing analysis, thus ruling out the Fah− cells with out-of-frame integrations at 5′ junction.
Importantly, the Fah−/− mice receiving Fah-MMEJ and Cas9 constructs showed reduced body weight loss, as compared to control Fah−/− mice receiving only Fah-MMEJ construct (Fig. 3e), indicating the corrected hepatocytes were functional in vivo and reduced the symptoms of liver damage. To further rescue the mice, after 3.5-week withdrawal of NTBC, the Fah−/− mice were re-administrated with NTBC for 4 days to allow liver recovery. During the second round of withdrawal of NTBC, the Fah-corrected hepatocytes continued to proliferate and repopulate the liver (Fig. 4a). Importantly, 5 of the 6 Fah-corrected mice survived, while all of the 6 control Fah−/− mice died within 9.5 weeks (Fig. 4b). Moreover, the Fah-corrected mice showed significantly reduced liver damage as indicated by decreased AST and ALT levels of the serum (Fig. 4c–d). These data demonstrated that the Fah−/− mice could be rescued by MMEJ-mediated correction of Fah gene.
Fig. 4.
MMEJ–mediated gene correction rescues lethal phenotype of HTI Fah-deficient mice. (a) Fah immunohistochemistry staining of liver sections from Fah−/− mice injected with MMEJ donor or MMEJ donor plus Cas9. Scale bar, 100 μm. (b) Survival rate of Fah−/− mice injected with MMEJ donor or MMEJ donor plus Cas9. Mice injected with MMEJ donor or MMEJ donor plus Cas9 in Fig. 3e were re-administrated with NTBC water for 4 days and then NTBC was withdrawn afterwards. MMEJ only, n = 6; MMEJ + Cas9, n = 6 mice. (c&d) Liver damage markers (aspartate aminotransferase (AST), alanine aminotransferase (ALT)) were measured in peripheral blood from Fah−/− mice injected with MMEJ donor only or MMEJ donor plus Cas9 after NTBC off 30 days. Fah−/− mice on NTBC water (NTBC on) served as a control. Results were presented as mean ± s.d. *p < 0.05, ***p < 0.001, unpaired Student's t-test.
4. Discussion
In this study, we found that MMEJ-mediated targeted integration showed much higher (up to 10 fold) knock-in efficiency than HDR-based strategy in adult mouse tissues. MMEJ is active during G1/early S phases, whereas HDR is active during late S/G2 phases. Difference in activity of MMEJ and HDR during the cell cycle may contribute to the higher efficiency of donors with short homology arms.
Recently, Zhang et al. reported that a double-cut donor vector with a long arm (400 bp) was superior to a long-arm donor without a double cut (HDR-based method) or a double-cut donor vector with a short arm (50 bp, MMEJ-based method) in human iPS cells. In contrast to human iPS cells (dividing cells), the majority of cells in vivo are non-dividing cells and thus MMEJ-based targeted integration may show more efficient than HDR-based targeted integration due to the cycle. Meanwhile, it is very interesting to investigate whether a double-cut donor vector with a long arm exhibit higher knock-in efficiency than a double-cut donor vector with a short arm in tissues in vivo.
Lately, non-homologous end joining (NHEJ)-based method was developed for efficient gene knock-ins in vivo (Suzuki et al., 2016). However, numerous studies (Kimura et al., 2014, Auer et al., 2014, Nakade et al., 2014, Hisano et al., 2015, Li et al., 2015, Cristea et al., 2013, Maresca et al., 2013) have shown that NHEJ repair system introduced random directions in integration and various types of indel mutations at the junctions, making it difficult to construct endogenous and exogenous fusion genes for chimeric proteins by in-frame integration. More importantly, NHEJ-based targeted integration can only introduce donor DNA segment into the cutting site, making it inapplicable to replace mutated sequence (such as point mutation, partial exon deletion or insertion, which are common mutation types in human diseases) with the correct one. By contrast, our MMEJ-based strategy introduced MMEJ-mediated integration (Kimura et al., 2014, Auer et al., 2014), a targeted integration in a homology-dependent manner, making DNA segment replacement in the genome practicable. Therefore, this MMEJ-based strategy may offer broader applications in gene therapy.
Funding Sources
This work was supported by CAS Strategic Priority Research Program (XDB02050007, XDA01010409), the MoST863 Program (2015AA020307), NSFC grants (31522037, 31500825), China Youth Thousand Talents Program (to H.Y.), Break through project of Chinese Academy of Sciences (to H.Y.). The Ministry of Science and Technology of China (MOST; 2016YFA0100500), the National Natural Science Foundation of China (NSFC; 31571509, 31522038), Shanghai Sailing Plan for the Young Scientific Talents (14YF1406900) (P.Y.H.)
Conflicts of Interest
The authors declare no competing financial interests.
Author Contributions
X.Y. and X.W. designed and performed experiments. J.L.L. and X.D.H. performed in vivo knock-in experiments. L.Y. S., X.W.S. and W.Q.Y. assisted with cloning and animal experiments. X.Y.S. and X.W. assisted with animal experiments. H.Y. and P.Y.H. supervised the project, designed experiments and wrote the manuscript.
Acknowledgement
We thank M.-M. Poo for comments on the manuscript.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2017.05.015.
Contributor Information
Pengyu Huang, Email: huangpy@shanghaitech.edu.cn.
Hui Yang, Email: huiyang@ion.ac.cn.
Appendix A. Supplementary data
Supplementary material
References
- Auer T.O., Duroure K., De Cian A., Concordet J.P., Del Bene F. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 2014;24:142–153. doi: 10.1101/gr.161638.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma H., Paulk N., Ranade A., Dorrell C., Al-Dhalimy M., Ellis E., Strom S., Kay M.A., Finegold M., Grompe M. Robust expansion of human hepatocytes in fah −/−/Rag2 −/−/Il2rg −/− mice. Nat. Biotechnol. 2007;25:903–910. doi: 10.1038/nbt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu V.T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., Kuhn R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- Cristea S., Freyvert Y., Santiago Y., Holmes M.C., Urnov F.D., Gregory P.D., Cost G.J. In vivo cleavage of transgene donors promotes nuclease-mediated targeted integration. Biotechnol. Bioeng. 2013;110:871–880. doi: 10.1002/bit.24733. [DOI] [PubMed] [Google Scholar]
- Grompe M., Al-Dhalimy M., Finegold M., OU C.N., Burlingame T., Kennaway N.G., Soriano P. Loss of fumarylacetoacetate hydrolase is responsible for the neonatal hepatic dysfunction phenotype of lethal albino mice. Genes Dev. 1993;7:2298–2307. doi: 10.1101/gad.7.12a.2298. [DOI] [PubMed] [Google Scholar]
- Hisano Y., Sakuma T., Nakade S., Ohga R., Ota S., Okamoto H., Yamamoto T., Kawahara A. Precise in-frame integration of exogenous DNA mediated by CRISPR/Cas9 system in zebrafish. Sci. Rep. 2015;5:8841. doi: 10.1038/srep08841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., He Z., Ji S., Sun H., Xiang D., Liu C., Hu Y., Wang X., Hui L. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- Kimura Y., Hisano Y., Kawahara A., Higashijima S. Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci. Rep. 2014;4:6545. doi: 10.1038/srep06545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor A.C., Badran A.H., Liu D.R. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell. 2017;168(1–2):20–36. doi: 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhang B.B., Ren Y.G., Gu S.Y., Xiang Y.H., Du J.L. Intron targeting-mediated and endogenous gene integrity-maintaining knockin in zebrafish using the CRISPR/Cas9 system. Cell Res. 2015;25:634–637. doi: 10.1038/cr.2015.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Miao Q., Yuan J., Han S., Zhang P., Li S., Rao Z., Zhao W., Ye Q., Geng J., Zhang X., Cheng L. Ascl1 converts dorsal midbrain astrocytes into functional neurons in vivo. J. Neurosci. 2015;35:9336–9355. doi: 10.1523/JNEUROSCI.3975-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca M., Lin V.G., Guo N., Yang Y. Obligate ligation-gated recombination (ObLiGaRe): custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Res. 2013;23:539–546. doi: 10.1101/gr.145441.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T., Dougan S.K., Truttmann M.C., Bilate A.M., Ingram J.R., Ploegh H.L. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakade S., Tsubota T., Sakane Y., Kume S., Sakamoto N., Obara M., Daimon T., Sezutsu H., Yamamoto T., Sakuma T., Suzuki K.T. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat. Commun. 2014;5:5560. doi: 10.1038/ncomms6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulk N.K., Wursthorn K., Wang Z., Finegold M.J., Kay M.A., Grompe M. Adeno-associated virus gene repair corrects a mouse model of hereditary tyrosinemia in vivo. Hepatology. 2010;51:1200–1208. doi: 10.1002/hep.23481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Tsunekawa Y., Hernandez-Benitez R., Wu J., Zhu J., Kim E.J., Hatanaka F., Yamamoto M., Araoka T., LI Z., Kurita M., Hishida T., LI M., Aizawa E., Guo S., Chen S., Goebl A., Soligalla R.D., Qu J., Jiang T., Fu X., Jafari M., Esteban C.R., Berggren W.T., Lajara J., Nunez-Delicado E., Guillen P., Campistol J.M., Matsuzaki F., Liu G.H., Magistretti P., Zhang K., Callaway E.M., Zhang K., Belmonte J.C. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540:144–149. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M., Nomura T., Osumi N. Transferring genes into cultured mammalian embryos by electroporation. Develop. Growth Differ. 2008;50:485–497. doi: 10.1111/j.1440-169X.2008.01046.x. [DOI] [PubMed] [Google Scholar]
- Yin H., Song C.Q., Dorkin J.R., Zhu L.J., Li Y., Wu Q., Park A., Yang J., Suresh S., Bizhanova A., Gupta A., Bolukbasi M.F., Walsh S., Bogorad R.L., Gao G., Weng Z., Dong Y., Koteliansky V., Wolfe S.A., Langer R., Xue W., Anderson D.G. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Xue W., Chen S., Bogorad R.L., Benedetti E., Grompe M., Koteliansky V., Sharp P.A., Jacks T., Anderson D.G. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 2014 doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Liu Y., Ma T., Liu K., Xu S., Zhang Y., Liu H., La Russa M., Xie M., Ding S., Qi L.S. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell. 2015;16:142–147. doi: 10.1016/j.stem.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material