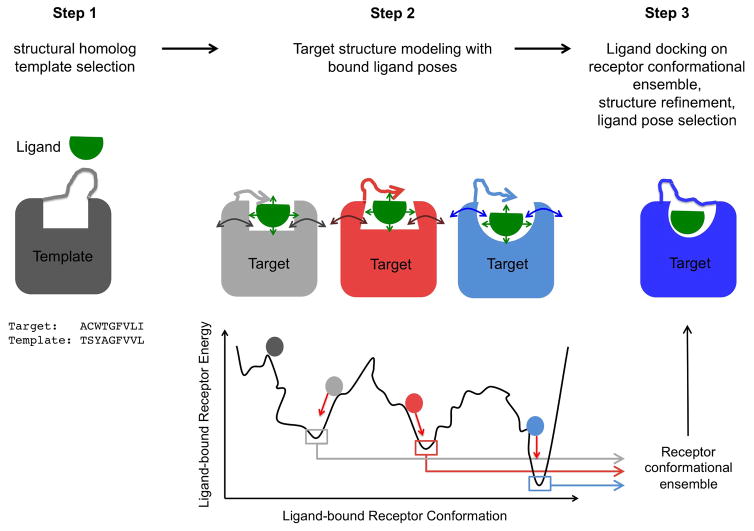
Figure 1. Integrated homology modeling and ligand docking.
Step1: generation of ligand conformers and selection of structural homolog template by sequence-structure alignment to the target sequence. Step2: target structure modeling in the presence of bound ligand involving simultaneous coarse-grained de novo reconstruction of receptor loops and sampling of ligand conformations followed by all-atom relaxation of ligand-bound receptor structures. Step3: All-atom docking of a large library of ligand conformers on the ensemble of low-energy receptor conformations generated in step 2 to refine the structure and the selection of receptor-bound ligand poses.