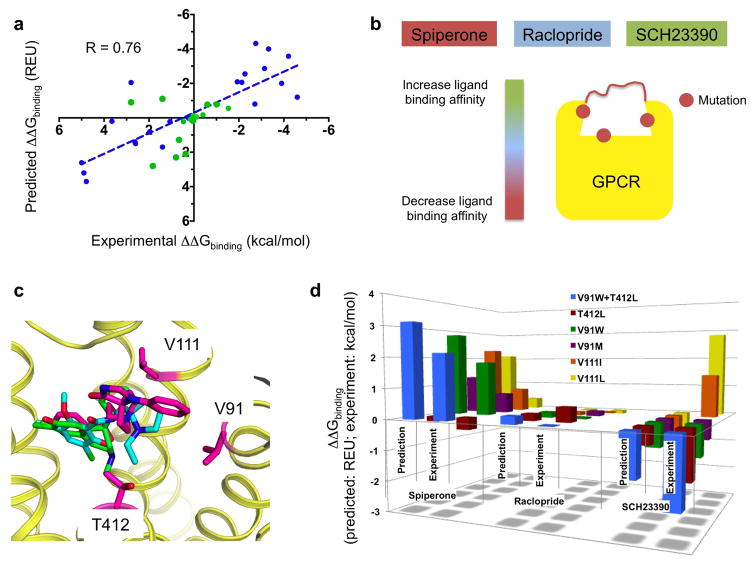
Figure 4. Prediction and design of ligand binding selectivity to structurally-uncharacterized GPCRs.
a. Correlation between predicted and experimentally measured ligand binding energy differences of a ligand for two receptors (blue: benchmark predictions with published experimental binding affinities; green: blind predictions). R: Pearson correlation coefficient (two-tailed P value < 0.0001). b. Schematic rationale for the structure-based design of the dopamine D2 receptor ligand binding site to reprogram the receptor’s ligand binding selectivity as follows: by simultaneously decreasing the binding to the high-affinity ligand spiperone, increasing the binding to the low-affinity ligand SCH23390 without affecting the affinity to the high-affinity ligand raclopride. c. Low-energy ligand poses bound to one conformation of the D2 receptor highlighting the positions of the designed specificity switch mutations (magenta: spiperone; cyan: raclopride; green: SCH23390). d. Comparison between blind predicted and experimentally-measured changes in binding energy of the 3 ligands for 6 designed D2 variants. Experimental values correspond to the mean of at least 3 independent experiments (see Methods, Supplementary Table 5). Each designed mutation has a significant effect on spiperone and SCH binding (all unpaired two-sided t-test P values < 0.05) but not on raclopride binding (all unpaired two-sided t-test P values > 0.1).