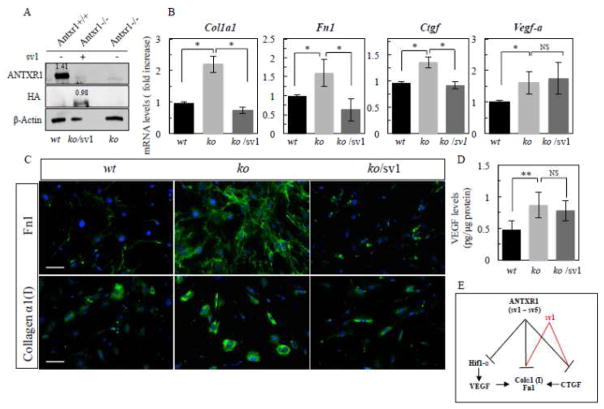
Fig. 5.
Re-expression of ANTXR1(sv1) restored levels of Col1a1 and Fn1 transcripts. (A) Assessment of sv1 transfection efficiency by western blotting of lysates from P49 control (wt) and Antxr1 null (ko) fibroblasts treated with vehicle or sv1pcDNA3-ANTXR1-sv1-HA (sv1) expression plasmid (ko/sv1). Numbers above the bands represent relative expression levels of ANTXR1 and ANTXR1-sv1 proteins normalized to β–actin. (B) Bar graphs showing average transcript levels (fold increase) of Ctgf, Col1a1, Fn1 and Vegf-a (*P<0.01; ns-not statistically significant, n = 4 per group). (C) Immunofluorescence staining for collagen α1(I) and fibronectin in cultures of fibroblasts from P49 control (wt) mice and fibroblasts from Antxr1 null (ko) mice treated with vehicle (ko) or sv1- containing expression plasmid (ko/sv1). Scale bar is 50 μm. Representative images are from triplicates of 2 independent experiments. (D) VEGF protein levels were assessed by ELISA in sv1 transfected and untransfected fibroblasts (**P<0.05; ns-not statistically significant, n = 4 per each group). (E) Schematic diagram of ANTXR1-dependent regulation of Col1a1 and Fn1 in fibroblasts. ANTXR1 (including all 5 splice variants sv1- sv5) negatively modulates Ctgf, Col1a1 and Fn1, Vegf-a and Hif1-α/expression (black lines) and sv1 negatively modulates Ctgf, Col1a1 and Fn1 only (red lines). ANTXR1-dependent increase of collagen α1(I) and fibronectin expression in mutant fibroblasts is primarily mediated by sv1/CTGF-dependent mechanism. Other ANTXR1 splice variants or unknown player (s), downstream of ANTXR1 may contribute to Hif1-α/VEGF-dependent matrix production in Antxr1 null fibroblasts.