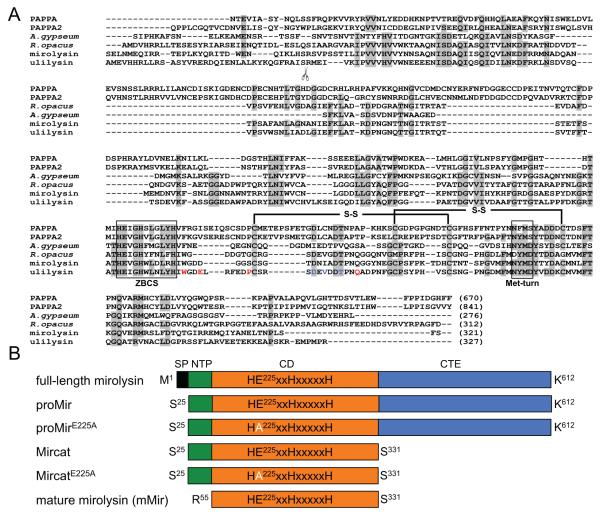
Figure 1.
Alignment of mirolysin with other members of the M43B protease family (A) and proteins used in this study (B). A) Sequence alignment of the catalytic domains of mirolysin (UniProt (UP) accession number: A0A0F7IPS1) and other pappalysins (family M43B) including human PAPPA (UP: Q13219) and PAPPA-2 (UP: Q9BXP8), ulilysin from Archaea (UP: Q8TL28), and proteases from the bacteria A. gypseum (UP: E5QZI4) and R. opacus (GenBank accession number: AII05686.1). Amino acid residues identical in a minimum of four out of six aligned sequenced are indicated by black letters on a gray background. Extended zinc-binding consensus sequences (ZBCS) and the Met-turn are boxed. Disulfide bonds identified for human pappalysin-1 (PAPPA) and ulilysin are marked with black lines. The scissors symbol indicates the site of autolytic cleavage during activation of ulilysin. The amino acid residues involved in the binding of two calcium ions by the protease are shown in red and blue fonts. B) Schematic drawing of the constructs and proteins expressed and purified in this study. SP – signal peptide; NTP – N-terminal profragment; CD – catalytic domain; CTE – C-terminal extension.